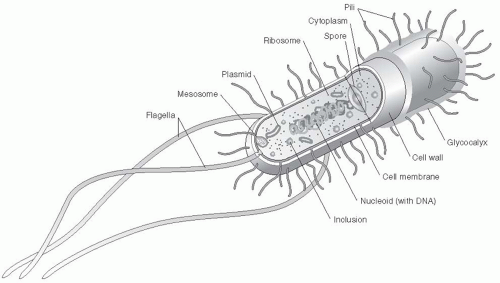
Organism |
Clinical Features/Disease |
Epidemiologic Features |
Aerobic and Facultatively Anaerobic Gram-Positive Cocci |
Staphylococcus aureus |
Cutaneous infections (e.g., abscess, impetigo, wounds); pneumonia; bacteremia; osteomyelitis; arthritis; toxic shock syndrome; food poisoning |
Colonize human skin and mucosal membranes (e.g., nares); resistance development (healthcare- and community-associated MRSA) |
Staphylococcus, coagulase negative (e.g., S. epidermidis) |
Opportunistic pathogen: bacteremia, endocarditis; prosthetic joint infects |
Colonize human skin and mucosal surfaces |
Enterococcus faecalis and E. faecium |
Bacteremia, endocarditis; urinary tract infections; intraabdominal abscesses |
Elderly patients; patients with long-term hospitalization and broad-spectrum antibiotic therapy |
Streptococcus pyogenes (group A) |
Pharyngitis; scarlet fever; sinusitis; impetigo, erysipelas, cellulitis, necrotizing fasciitis; glomerulonephritis; rheumatic fever; toxic shock syndrome |
Diverse populations; carrier state |
Streptococcus agalactiae(group B) |
Neonatal disease (bacteremia, meningitis, pneumonia); urinary tract infections in adults |
Diverse populations; carrier state |
Viridans group streptococci |
Abscess formation; subacute endocarditis; dental caries |
Patients with abnormal or prosthetic heart valves |
Streptococcus pneumoniae |
Pneumonia and other respiratory tract infections; meningitis; bacteremia; endocarditis |
Diverse populations: neonates, children, adults with chronic pulmonary disease, elderly |
Aerobic or Facultatively Anaerobic Gram-Positive Rods |
Bacillus anthracis |
Anthrax (cutaneous, inhalational, gastrointestinal) |
Animal workers; laboratory accidents; bioterrorism; consumption of infected meat |
Bacillus cereus |
Gastroenteritis, bacteremia, ocular infections |
Contaminated food; traumatic eye injury; IV drug use |
Corynebacterium diphtheriae |
Diphtheria (respiratory or cutaneous) |
Spread by respiratory droplets |
Erysipelothrix rhusiopathiae |
Erysipeloid (cellulitis) |
Occupational disease (e.g., butchers, meat processors, farmers) |
Listeria monocytogenes |
Early-onset and late-onset neonatal disease (meningitis, sepsis); bacteremia in pregnant women |
Ingestion of contaminated food (nonpasteurized dairy products); immunocompromised hosts; elderly women |
Mycobacteria |
Mycobacterium tuberculosis |
Tuberculosis |
Opportunistic pathogen; emerging multidrug resistance |
Mycobacterium avium complex |
Localized pulmonary disease; also disseminated disease with multiorgan disease |
Localized in patients with chronic pulmonary disease; disseminated in patients with HIV/AIDS |
Mycobacterium leprae |
Leprosy (Hansen’s disease) |
Spread by close contact with infected patients |
Other Gram-Positive, Partially Acid-Fast Bacteria |
Nocardia species |
Bronchopulmonary disease |
Commonly found in soil and water; opportunistic pathogen in patients with underlying pulmonary disease |
Rhodococcus equi |
Lung abscess; opportunistic infections in immunocompromised patients |
Commonly found in immunocompromised patients (e.g., AIDS, transplant recipients) |
Aerobic Gram-Negative Cocci |
Neisseria meningitides |
Meningitis, bacteremia |
Carrier state; aerosol transmission |
Neisseria gonorrheae |
Gonorrhea, pelvic inflammatory disease, arthritis |
Sexual transmission; asymptomatic carriers |
Aerobic and Facultatively Anaerobic Gram-Negative Rods |
Escherichia coli |
Urinary tract infections; bacteremia; meningitis |
UTI: sexually active women Meningitis: neonates |
Escherichia coli, enterotoxic (STEC), enterohemorrhagic, and entoaggregative strains |
Watery diarrhea, hemorrhagic colitis; watery diarrhea with mucus; hemolytic-uremic syndrome (HUS) |
Foodborne or waterborne outbreaks; travelers’ diarrhea; infants in developing countries |
Klebsiella pneumonia Serratia marcescens Enterobacter species |
Pneumonia; urinary tract infections Pneumonia; urinary tract infections; wound infections |
Nosocomial infections; alcoholism Nosocomial infections |
Proteus species |
Urinary tract infections; wound infections |
Structural abnormalities in the urinary tract |
Vibrio cholera |
Cholera; severe watery diarrhea |
Children and adults in developing countries |
Vibrio vulnificus |
Wound infections; primary septicemia |
Immunocompromised patients; preexisting (chronic) hepatic disease |
Vibrio parahaemolyticus |
Watery diarrhea |
Seafood-borne outbreaks |
Aerobic and Facultatively Anaerobic Gram-Negative Rods |
Salmonella species |
Diarrhea; enteric fever (serovar typhi) |
Contaminated food; immunocompromised patients are at risk for bacteremia |
Shigella species |
Bacillary dysentery |
Contaminated food or water; person-to-person transmission |
Yersinia pestis |
Plague |
Natural reservoir: rodents; transmitted to humans by rodent flea bites; bioterrorism |
Yersinia enterocolitica and Y. pseudotuberculosis |
Enterocolitis; mesenteric lymphadenitis |
Worldwide distribution; contaminated food and water |
Aeromonas species |
Wound infections; gastroenteritis |
Widely distributed in aquatic reservoirs; contaminated drinking water |
Acinetobacter baumaniicomplex |
Pneumonia; septicemia; wound infections; opportunistic infections |
Nosocomial infections; patients with large wounds or burns are at increased risk; elderly |
Pseudomonas aeruginosa |
Pulmonary infections; primary skin infections; infections of urinary tract, ear, eye, and wounds; bacteremia |
Nosocomial infections; ubiquitous organism in nature, in aquatic reservoirs, and on surfaces |
Stenotrophomonas maltophilia |
Wide variety of serious disseminated infections |
Nosocomial infections |
Burkholderia cepacia complex |
Pulmonary infections; opportunistic infections |
Compromised patients, especially those with cystic fibrosis and chronic granulomatous disease |
Burkholdreia pseudomallei |
Melioidosis |
Opportunistic pathogen |
Moraxella catarrhalis |
Ear, eye, and respiratory tract infections |
Children; patients with underlying pulmonary disease |
Fastidious and Other Gram-Negative Bacilli |
Eikinella corrodens |
Subacute endocarditis; wound infections |
Human bite wounds; opportunistic pathogen in patients with damaged heart valves |
Kingella kingae |
Subacute endocarditis |
Opportunistic pathogen |
Haemophilus influenzae |
Encapsulated type b strains: meningitis; septicemia; epiglottitis Nonencapsulated strains: otitis media; sinusitis; bronchitis; pneumonia |
Aerosol transmission in young and unimmunized patients; possible spread from upper respiratory tract of elderly patients with chronic respiratory disease |
Legionella pneumophilia |
Legionnaires’ disease (pneumonia) and Pontiac fever (flu-like illness) |
Waterborne transmission; elderly and immunocompromised patients at increased risk |
Francisella tularensis |
Tularemia (pulmonary or skin infections) |
Human infections via contact with wild rabbits, ticks, or deerflies; bioterrorism |
Fastidious and Other Gram-Negative Bacilli |
Bordetella pertussis |
Pertussis (whooping cough) |
Aerosol transmission; pertussis toxin; severe disease in infants, milder in adults |
Brucella species |
Brucellosis |
Exposure to infected goats, sheep, and cattle |
Pasteurella multocida |
Wound infections; pulmonary and disseminated infections |
Cat bites; commensal in upper respiratory tract of animals, fowl, and perhaps humans |
Campylobacter jejuni |
Gastroenteritis |
Contaminated food, milk, or water |
Helicobacter pylori |
Gastritis; peptic and duodenal ulcers; gastric adenocarcinoma |
Infections are common (people in lower socioeconomic classes and in developing countries) |
Anaerobic Gram-Positive Organisms |
Clostridium perfringens |
Cellulitis, necrotizing fasciitis, myonecrosis; food poisoning; septicemia |
Ubiquitous in environment (e.g., soil, water, sewage) |
Clostridium difficile |
Antibiotic-associated diarrheal disease; pseudomembranous colitis |
Colonizes human GI tract; nosocomial, hospital pathogen |
Clostridium tetani |
Tetanus |
Ubiquitous in environment (e.g., soil, water, sewage) |
Clostridium botulinum |
Botulism (foodborne, infant, wound type) |
Ubiquitous in environment (e.g., soil, water, sewage) and GI tract of animals; bioterrorism |
Anaerobic Gram-Negative Organisms |
Bacteroides fragilis |
Polymicrobial infection of the abdomen, skin, soft tissue, and female genitourinary tract |
Colonizes human mucosal surface (oropharynx, intestine, vagina) |
Spirochetes |
Treponema pallidum |
Syphilis |
Worldwide distribution; sexually transmitted disease |
Borrelia burgdorferi |
Lyme disease |
Reservoir: rodents, deer, domestic pets, hard-shelled ticks Vector: hard-shelled ticks |
Borrelia recurrentis |
Epidemic, louse-borne relapsing fever |
Reservoir: humans Vector: body louse |
Leptospira |
Leptospirosis (asymptomatic to influenza-like illness with yalgia); Weil syndrome (jaundice) |
Worldwide distribution; rodents are common reservoir; soil is contaminated by rodent urine |
Mycoplasma and Gram-Negative, Obligate Intracellular Bacteria |
Mycoplasma pneumoniae |
Tracheobronchitis, pharyngitis, pneumonia |
Strict human pathogen; asymptomatic carrier state |
Chlamydophila pneumoniae |
Asymptomatic respiratory tract infections to “atypical” pneumonia |
Human pathogen |
Chlamydophila psitacci |
Psittacosis/ornithosis; pulmonary disease with hematogenous dissemination to liver and spleen |
Exposure to dried excrement, urine, or secretions from psittacine birds (parrots, parakeets, macaws, cockatiels) |
Chlamydia trachomatis |
Trachoma and lymphogranuloma venerum (LVG) |
Direct transmission by droplets via hands, eye-to-eye, or contaminated clothing; sexually transmitted |
Ehrlichia chaffiensis |
Human monocytic ehrlichiosis |
Region: southeastern, central, and midwestern United States Reservoir: white-tailed deer, foxes, coyotes, wolves, domestic dogs Vector: Lone Star tick (Amblyomma americanum) |
Anaplasma phagocytophilum |
Human granulocytic ehrlichiosis |
Region: northern United States central Midwest Reservoir: small mammals (mouse, chipmunks, voles) Vector: Lone Star tick and Ixodes tick |
Rickettsia prowazekii |
Epidemic typhus |
Human body louse (Pediculus humanus) More than 90% of infections in the United |
Rickettsia rickettsia |
Rocky Mountain spotted fever |
States occur from April to September Vector: dog tick (Dermacentor variabilis) and wood tick (Dermacentor andersoni) |
Coxiella burnetii |
Asymptomatic to flu-like symptoms; also pneumonia and hepatitis |
Wide variety of hosts: cattle, sheep, goats, dogs, cats, and rabbits; exposure occurs via contaminated soil and milk |