Table 15-1 The Genes of Influenza A Virus and Their Protein Products | |||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
from the host epithelial cell; specific anti-NA antibody presumably diminishes release of virons from host cells.
Table 15-2 Antigenic Shifts of Influenza A Virus | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||
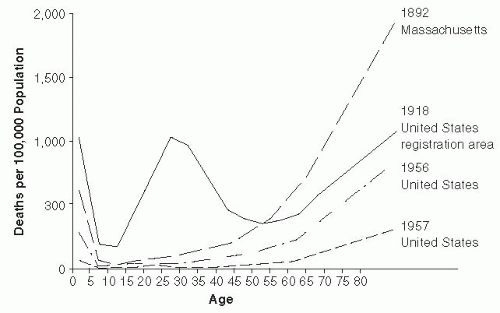 Figure 15-1 Age Distribution of Mortality of Selected Influenza Epidemics in the United States. Reproduced from C.C. Dauer and R.E. Sterling, 1961, Mortality from Influenza, American Review of Respiratory Diseases, vol. 82, Supplement, pp. 15-26. Official Journal of the American Thoracic Society. |
of 1918-1919. Philadelphia reported the highest mortality rate: 12,897 influenza and pneumonia deaths in October and November of 1918, with a peak of 700 deaths per day in late October, and a 2-month mortality rate of 0.77%, leading to disruption of civic life, including a shortage of coffins. Desperate medical and public health authorities recommended many remedies and preventive actions now regarded as ineffective, including the use of gauze face masks, aerosol sprays, garlic or camphor necklaces, and legislation against public spitting. Mortality rates were lower in military and civilian African Americans than in whites, but approximately 2% of all Native Americans died during the epidemic.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree








