FIGURE 2-1. Chemical structure of DNA and its relationship to chromatin. The most basic unit of the chromosome is DNA, which is composed of polynucleotide chains made up of four different nucleobases: thymine, adenine, cytosine, and guanine. Each polynucleotide chain runs in opposite directions to form a right-handed double helical structure with hydrogen bonding between complementary base pairs, where adenine always pairs with thymine, and guanine always pairs with cytosine. This double helix is compacted by wrapping around a protein octamer consisting of two copies each of four core histone proteins (H3, H4, H2A, and H2B) forming a nucleosome. Nucleosomes are further condensed via another histone (H1), which links flanking DNA that enters and leaves the core particle and functions to pack nucleosomes on each other to form solenoid structures.
Although chromatin is essential for the compaction of the eukaryotic genome, it creates a formidable obstacle between the gene-expression machinery and DNA regulatory elements. In fact, the topologic problem becomes even more complex as the “beads on a string” become more compacted. Histone H1 proteins are responsible for packing nucleosomes on each other to form solenoid structures (see Fig. 2-1),2,4,7 higher-order arrays in which DNA is further condensed to form euchromatin. Heterochromatin is the highly compacted form of chromatin that makes DNA sequences structurally inaccessible to the transcription machinery, resulting in functionally inactive genes. In fact, chromosomes represent the culminating form of compaction. These are transcriptionally inactive, transient structures that occur only during a unique temporal period of the cell cycle that leads ultimately to DNA replication and cell division.10
In sum, nucleosomes may be randomly or very specifically located over the bulk of chromosomal DNA and provide an important conceptual framework for fully understanding how hormones regulate gene transcription. The structure of chromatin is dynamic, with the state of the nucleosome core playing a pivotal role in governing the transcriptional competence of the targeted genes. Consequently, acetylation (associated with activation) and deacetylation (associated with repression), as well as methylation (associated with both activation and repression depending on the residue modified) of histone proteins, represent important steps that must be accommodated in a mechanistic model that defines hormone action.4 We will explore this issue in greater detail later in the chapter (see the section entitled “Hormonal Control of Gene Expression and Protein Biosynthesis”).
Functional Anatomy of a Gene
Within the vast amount of DNA from each eukaryotic cell, it is currently estimated that there are approximately 20,000 to 25,000 protein-coding genes in the human genome. Although Gregor Mendel called them particulate factors instead of genes in 1865,1 he clearly characterized their essential attributes. Strictly defined, a gene is the region of DNA transcribed by RNA polymerase.1,4 Regions of the transcribed gene found in mature mRNA are referred to as exons, short for expressed regions of DNA (Fig. 2-2). The precursor hnRNA exons are interrupted by intervening sequences (introns) that are excised as the nascent transcript is processed to its mature form. Steps involved in RNA processing are explored in more detail below.
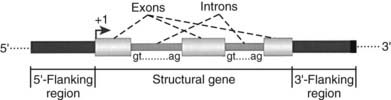
FIGURE 2-2. Functional anatomy of a gene. Regions of the structural gene that are retained in mature mRNA are known as exons, while the intervening sequences that are excised are called introns. The 5′ and 3′ sequences of all introns are conserved, encoding the splice donor (gt) and splice acceptor (ag), respectively. The region immediately upstream of the first transcribed nucleotide is referred to as the 5′-flanking region, and the portion of the gene that is located downstream of the structural gene is referred to as the 3′-flanking region. The gene promoter is typically located in the 5′-flanking region, allowing for the correct initiation and efficiency of transcription. The nucleotide where transcription begins is designated +1.
The region immediately upstream of the first transcribed nucleotide is referred to as the 5′-flanking region (see Fig. 2-2).1,2,4 Within this region lies the promoter, which contains all the information necessary for specifying the correct initiation of transcription and regulates the efficiency of transcription. Typically, the nucleotide where transcription begins is designated +1. Consequently, most portions of a promoter are denoted by a negative numbering system of nucleotides, indicating the upstream positioning of the domain. Achieving accurate initiation of transcription is essential for ensuring constancy of the reading frame used for translation of the transcribed mRNA. Modulating the efficiency of transcription gives cells the capacity to produce more or less protein as the need arises.
Since a gene is typically defined as the region transcribed by RNA polymerase, all transcribed regions downstream of the +1 nucleotide fall within this functional domain. Most genes encoding mRNA (those transcribed by RNA polymerase II) begin with a purine, either A or G (Fig. 2-3). However, defining the end of a gene transcribed by RNA polymerase II is more problematic. Unlike prokaryotic genes, there is no fixed site that specifies termination of transcription.1 Instead, a posttranscriptional processing event, addition of a homopolymeric tail of adenine nucleotides (poly A) signifies the end of the precursor hnRNA that will be further processed to generate mature mRNA.1–411 The enzyme that specifies polyadenylation, poly A polymerase, recognizes a specific hexameric sequence (AATAAA) and then cleaves the precursor mRNA approximately 29 bp downstream, with the resulting 3′-OH group used as the substrate for subsequent addition of approximately 200 to 250 adenine residues.1,3,11 The region of the gene that extends beyond the site of polyadenylation is referred to as the 3′-flanking region.
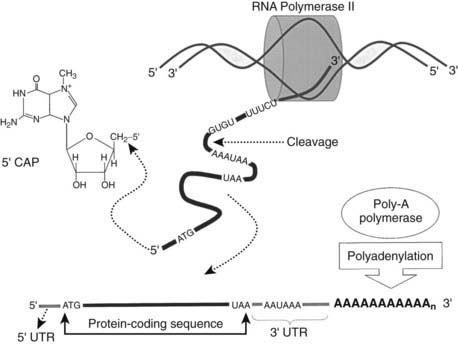
FIGURE 2-3. Transcription by RNA polymerase II creates the template for protein synthesis. Messenger RNA is the single-stranded molecule that transfers the genetic information from DNA in the nucleus to the cytoplasm, where proteins are translated. Mature mRNA is “capped” by addition of 7-methylguanosine to the 5′ end through a triphosphate linkage formed between its 5′-hydroxyl and the 5′-hydroxyl of the terminal residue in the untranslated region (5′ UTR) of the initial transcript. The 3′ ends of growing transcripts are cleaved between the polyadenylation sequence and sequences rich in guanine and uracil found in the 3′ untranslated region (3′ UTR). Following this cleavage event, poly-A polymerase enzyme adds 200 to 250 adenine residues. Both modifications of the mRNA confer mRNA stability, translational efficiency, and play a role in exportation of the mature mRNA from the nucleus to the cytoplasm.
In most mRNAs transcribed by RNA polymerase II, the start codon that specifies the beginning of the translation reading frame (ATG) is located between 5 and 100 bp downstream from the 5′-end of the transcribed mRNA.1,3 Thus the region between the 5′-end of the mRNA and the translation start site is referred to as the 5′-untranslated region. Similarly, the codon that defines the end of the translation reading frame (UAG, UAA, or UGA) is usually followed by a relatively long run of nucleotides before reaching the hexanucleotide sequence that defines the site for polyadenylation (see Fig. 2-3). This region is referred to as the 3′-untranslated region.
The processing of the precursor mRNA (hnRNA) will be described in more detail in a subsequent section (see the section entitled “Posttranscriptional Modification of mRNA”). However, before we treat this subject, it is useful to note that the exact functional significance of introns remains unclear. There are cases where microRNAs (miRNAs) are produced from the intronic region of a gene (as discussed in section entitled “RNAi Regulation of Gene Expression”). In other cases, some mRNAs can undergo alternative processing. When this occurs, a specific transcribed segment can either be retained and act as an exon or can be excised and act as an intron. Introns that can act as exons when retained contain long open reading frames that encode a polypeptide fragment. The unique duality of this type of intron may allow for the shuffling of functional units to create families of related products from a single gene.1,3
Functional Anatomy of the Promoter-Regulatory Region
In general, a promoter contains two functional domains. The core region of the promoter is defined as the minimal 5′-flanking region that is required for accurate initiation of transcription. The second promoter domain usually resides immediately upstream and contains one to several regulatory elements that regulate the level of transcription in various cell types and in response to extracellular signals. Since these elements are physically linked to the gene they regulate, they are referred to as cis–acting regulatory elements. However, the functionality of these elements emerges only on the binding of a specific transcription factor that is almost always encoded by a different gene. Hence, cis-acting elements bind trans-acting factors.
Transcription factors are modular and contain at least two functional domains: one that binds specifically to a given cis-acting element and one that directly or indirectly either influences correct initiation or modulates the efficiency of transcription. The boundaries of cis-acting elements are defined by the region of DNA that is actually contacted by the DNA-binding domain of a specific trans-acting factor and are usually less than 20 bp in length.1,2 Frequently they contain a core recognition sequence of 8 to 10 bp that is often palindromic,1,2 reflecting twofold symmetry of transcription-factor binding as dimers composed of the same subunit (homodimers) or different subunits (heterodimers).
The core promoter usually consists of an initiator element (Inr) that encompasses the transcription start site and a TATA box that is typically located 25 to 35 bp upstream of the transcription start site in higher eukaryotes and binds TATA-binding protein (TBP) (Fig. 2-4).12,13 TBP is a key component of transcription factor (TF) IID, a general transcription factor that binds DNA in a sequence-specific manner.3,4,12 TBP binds in the minor groove of the DNA double helix and forms the foundation of the preinitiation complex. Native TFIID is a large multi-subunit protein (>700 kD) consisting of TBP and at least eight TATA-associated factors (TAFs).12,13
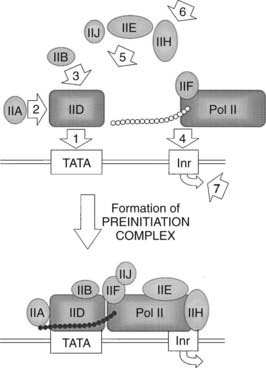
FIGURE 2-4. Assembly of the basal transcription machinery. The first step in formation of the preinitiation complex is the recognition and binding of TFIID (TBP plus 8 TAFs) to the TATA box. The second step consists of the coupling of TFIIA to TFIID, stimulating and stabilizing TFIID binding. The third step involves TFIIB binding to either TFIID or the TFIID/TFIIA complex. Fourth is the association of the unphosphorylated form of RNA polymerase II (Pol IIA) with the growing complex. The fifth step consists of the sequential binding of TFIIE, TFIIH, and TFIIJ to form the preinitiation complex. The sixth step involves the enzymatic activities of TFIIH allowing the phosphorylation of RNA polymerase II (Pol IIO), melting of the DNA duplex at the transcription start site, and the release of TFIIE, TFIIB, and two subunits of TFIIH. Finally, TFIIA and TFIID remain bound to the promoter while Pol IIO, TFIIF, and one subunit of TFIIH move to form the elongation complex.
Once TFIID binds, several other general transcription factors follow in an ordered succession, forming an extremely large core transcription complex (see Fig. 2-4). TFIIA, composed of three subunits (14, 19, and 34 kD), binds TFIID and DNA upstream of TBP, although this event is not DNA sequence-specific.2,4,12 TFIIA stabilizes TFIID and causes a conformational change in TBP that may displace a negative component in the native TFIID.12 TFIIB, a single 35-kD subunit, binds to and stabilizes the TFIID-IIA complex. TFIIF, consisting of two polypeptides (30 and 74 kD), forms a molecular bridge with TFIIB between RNA polymerase II (Pol II) and TBP.4,12 Both TFIIB and TFIIF appear to function in start-site selection. RNA polymerase II consists of 10 polypeptides, ranging in size from 10 to 240 kD, the largest of which contains an unusual C-terminal domain (CTD) that is extensively phosphorylated.12,14 The unphosphorylated form of Pol II (Pol IIA) preferentially associates with the committed complex relative to the phosphorylated form (Pol IIO). TFIIE, which functions as a tetramer (two copies each of 34 and 56 kD subunits), binds TBP, TFIIF, Pol IIA, and TFIIH, the next protein to bind the growing complex.15 TFIIH (at least eight subunits totaling 200 to 300 kD) is the only general transcription factor to show catalytic activity, including CTD kinase activity that is regulated by TFIIE.12,15 Additionally, TFIIH appears to function as a helicase and a DNA-dependent ATPase.12,15 TFIIJ is the last factor to enter the preinitiation complex. Although it is known that TFIIJ is required, the function of this factor has not been characterized. It is the formation of this core transcription complex that determines the accurate initiation of transcription.
A newer player on the transcriptional scene is a complex called the Mediator. This complex is composed of up to 30 subunits in mammals and has been found to be involved with formation of the preinitiation complex. It can function in the basal expression of genes, as well as in both activator-dependent transcription and the repression of transcription. Mediator is as critical for transcription as polymerase II itself. It binds to the unphosphorylated carboxyl terminal domain (CTD) of the largest subunit of RNA polymerase II and is thought to be involved during assembly of the preinitiation complex, either during the recruitment of Pol II, TFIID, and other general transcription factors or by increasing the efficiency and rate of the preinitiation complex. Mediator disassociates from RNA polymerase II when the CTD becomes hyperphosphorylated, upon the start of transcriptional elongation. There are four distinct modules that make up the Mediator complex: the head, middle, tail, and CDK. It appears there is a core complex that enables additional components to be added to allow for cell-specific changes to external signals.16 Mediator also has a functional role in chromatin remodeling. It has been shown to be involved in gene silencing of neuronal genes in extraneuronal cells by interaction with G9a histone methyl transferase and REST (RE1 silencing transcription factor).17 Transcriptional initiation requires not only the core transcription complex but also the mediator complex and nucleosome-modifying enzymes.
Although TFIID is capable of recognizing several nonconsensus TATA sequences, some promoters clearly lack a TATA box.2,18 This is especially true for promoters in some housekeeping genes such as the gene encoding an enzyme that catalyzes the formation of adenosine monophosphate from adenine and phosphoribosylpyrophosphate.1 This protein acts as a salvage enzyme for recycling of adenine into nucleic acids. Preinitiation complex assembly on these TATA-less promoters is mediated through the Inr, the consensus sequence of which is pyrimidine-pyrimidine-A-N-T/A-pyrimidine-pyrimidine, A being the transcription start site at 1.18 In these cases, Pol II recognizes and binds the Inr directly and nucleates the binding of the other factors in the preinitiation complex.19
One to several cis-acting elements are located in close proximity and 5′ to the TATA box (Fig. 2-5). These accessory elements set the basal transcriptional tone of the promoter by increasing the efficiency of transcription. The trans-acting factors that bind these elements are generally ubiquitous, including Sp1 and NF-Y, which bind GC-rich regions and CCAAT boxes, respectively.1,2,20 The binding of these factors to DNA results in protein-protein interactions with the basal transcription machinery to increase or decrease transcription in a non-tissue-specific manner. Given the ubiquitous presence of factors such as Sp1 and NF-Y, it is not surprising that their corresponding cis elements are located on promoters of many genes, including housekeeping genes that provide basic functions needed for maintenance of all cell types.
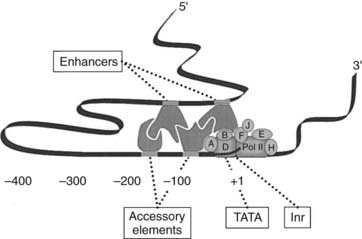
FIGURE 2-5. Functional anatomy of the promoter-regulatory region. The core region of the promoter, defined as the minimal 5′-flanking region required for accurate initiation of transcription, is typically made up of the initiator element (Inr), which encompasses the transcription start site, and a TATA box. The second domain resides in close proximity to the core promoter and contains one to several accessory elements that modulate the efficiency of transcription. Enhancers are another class of promoter regulatory elements that are usually located further upstream from the gene to which they regulate. Contact between trans-acting factors that bind enhancers, accessory elements, and the basal transcription machinery occurs through looping of the DNA. All transcription factors that bind regulatory elements contain a domain that binds specifically to a given cis-acting element and another domain that directly or indirectly influences transcription.
Given their close proximity and direct interaction with the core transcriptional machinery, accessory elements are position and orientation dependent. This is in contrast to enhancers, another class of promoter regulatory elements (see Fig. 2-5), which are located farther upstream from 100 bp to several thousand bp or even 3′ to the gene they regulate.1–4 When assayed for activity by attachment to a heterologous core promoter, activities of enhancers display considerable distance, orientation, and position independence. Nevertheless, the trans-acting factors that bind this class of regulatory elements must also make contact with the core transcriptional machinery. Although the distance between an enhancer and TATA box may be considerable, this contact may occur through looping of the DNA.1–3
Enhancers represent a broad class of elements that are capable of binding a variety of transcriptional factors. Some of these are tissue or cell specific and thus confer this property to the promoter they regulate. In addition to increasing transcription, enhancers can also repress transcription, depending on the nature of the protein they bind. While some enhancers act alone, others are represented by tightly packed arrays of cis-acting elements and are designated as composite enhancers.1 In fact, it is not uncommon to find that tissue- or cell-specific expression is determined by the concerted action of composite enhancers that bind both ubiquitous and tissue- or cell-specific proteins.
In addition to housing elements that determine basal transcriptional tone and spatially restricted expression, promoter regulatory regions also contain cis-acting elements that confer responsiveness to a wide variety of homeostatic agents, such as hormones, and to an equally wide array of environmental cues and insults. These elements are referred to as response elements. Like the elements noted above, response elements can bind proteins that are either ubiquitous or relatively cell-type specific. One such inducible factor, presumably activated by stress, is the heat shock transcription factor (HSTF) that binds heat shock elements (HSEs).1 Normally, this factor exists in cells but is inactive. When cells are insulted by a sudden increase in temperature, HSTF becomes active and binds HSEs located in the promoters of genes that encode proteins. This aids in cell survival at higher temperatures. An explanation about how hormones bind response elements and either induce or repress transcription of the genes they regulate will be explored later (see the section entitled “Hormonal Control of Gene Expression”).
Transcription: Creating the Template for Protein Synthesis
Although DNA is the genetic material, it does not function as the scaffold for protein synthesis. Messenger RNA is the single-stranded intermediate molecule that transfers the genetic information from DNA in the nucleus to the cytoplasm, where it serves as a template in the formation of polypeptides. RNA is quite similar in structure to DNA; in fact, a single strand of RNA can even form a double-stranded hybrid helix with a DNA strand. One minor difference between RNA and DNA involves the pentose sugar of RNA.1,2 It contains an additional hydroxyl group (ribose as opposed to deoxyribose). In addition, uracil (U) replaces T in RNA. Despite these subtle differences, organisms have evolved mechanisms allowing for a smooth transition from DNA to RNA through transcription.
Transcription is the first step in which genetic information is converted from DNA into RNA and proteins. It is also the major point at which gene expression is regulated. A eukaryotic gene can be classified on the basis of the enzyme that drives its transcription. RNA polymerases are multi-subunit enzymes that synthesize RNA using a DNA template. The most active of the RNA polymerases is RNA polymerase I, which resides in the nucleolus and is responsible for transcribing genes encoding ribosomal RNA (rRNA), a major component of ribosomes.1,3,4 RNA polymerase II, or Pol II, as was mentioned previously, is also a highly active nuclear enzyme that is responsible for synthesizing hnRNA, a precursor to mRNA.1,3,4 The final RNA polymerase, RNA polymerase III, transcribes transfer RNA (tRNA), an adapter molecule involved in translation. This chapter will remain focused on genes whose expression is transcribed by Pol II.
Posttranscriptional Modification of mRNA
During synthesis, immature mRNAs are covalently modified at both their 5′ and 3′ ends (see Fig. 2-3). Almost immediately following initiation of precursor mRNA synthesis, the 5′ end of the molecule is “capped” by addition of a methylated guanosine.1,3,4 7-Methylguanosine is attached through a triphosphate linkage formed between its 5′-hydroxyl and the 5′-hydroxyl of the terminal residue in the initial transcript. This cap plays a role in nuclear transport of the mRNA. Additionally, the 5′ cap is essential for most mRNA translation because it facilitates binding of the translation machinery to the 5′ end of the mRNA.1,3 This modification also protects the fragile mRNA from degradation as the unique 5′ to 5′ phosphodiester bond of the cap makes it intrinsically resistant to general ribonucleases.21
The 3′ ends of growing transcripts are cleaved at a point 10 to 30 bases downstream of the polyadenylation signal sequence, AAUAAA (see Fig. 2-3). This sequence is found in nearly all eukaryotic mRNAs and is one of the most conserved elements known.1,3,4,11,22 Other elements, containing GUGU and UUUCU sequences, are located 20 to 40 bases downstream of the cleavage site. Immediately following cleavage of the nascent transcript, poly-A polymerase enzyme adds 200 to 250 adenylate residues.1,22 Like the 5′ cap, this modification confers mRNA stability, promotes mRNA translational efficiency, and plays a role in mature mRNA export from the nucleus to the cytoplasm.1,3,22
Many mRNA precursors in the nucleus are much larger than their cytoplasmic mRNA counterparts associated with ribosomes. Excision of the intronic, or noncoding, sequences (Fig. 2-6) is the most significant modification that mRNA undergoes before the mature form is transported to the cytoplasm. Each intron contains conserved sequences at the 5′ and 3′ ends, known as the splice donor (GU) and acceptor (AG), respectively.1,3,4 An array of small ribonucleoproteins and associated nuclear proteins form a complex known as the spliceosome, which recognizes the ends of the intron and brings them together.1,3,4 The immature mRNA is cleaved immediately upstream of the splice donor at the 5′ end of the intron, and the terminal G covalently links to an A found near a pyrimidine-rich region that precedes the splice acceptor, forming a lariat structure.1,3,4 The lariat is cleaved immediately downstream of the splice acceptor, and the intron is rapidly degraded while the adjacent two exons are joined together.
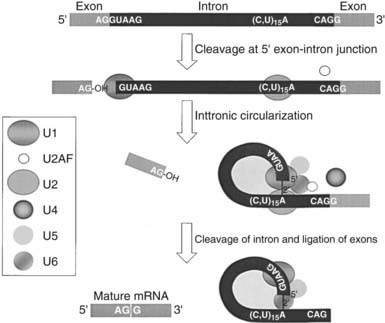
FIGURE 2-6. Splicing is a posttranscriptional modification of mRNA. Splicing involves the excision of intronic sequences from the mRNA before the mature form is transported to the cytoplasm. Immature mRNA is cleaved immediately upstream of the splice donor (GU) at the 5′ end of the intron, and the terminal G nucleotide covalently links to an A residue found near a pyrimidine-rich region near the 3′ end of the intron, forming a lariat structure. A large array of small ribonucleoproteins and associated nuclear proteins identified in the box to the left form a complex known as the spliceosome, which recognizes the ends of the intron and brings them together. This lariat is cleaved immediately downstream of the splice acceptor (AG), and the adjacent two exons are joined together while the intron is degraded.
Alternative splicing of precursor mRNAs is a common mechanism whereby cells exploit the splicing mechanism to generate multiple related proteins from a single gene.23 Once thought to be an exception to the rule (one gene, one protein), alternative splicing is now estimated to occur in at least 1 of every 20 genes.23 One example of a single gene that is alternatively spliced is α-tropomyosin, which encodes seven tissue-specific variants of the muscle protein that associates with actin in the rat.3,4 This gene consists of “constitutive” exons that are found in all transcripts of the gene, “cell-specific” exons that appear only in transcripts produced in certain tissues, and exons that show variable expression. The mechanism of splice site selection and the interaction between multiple cis-acting elements and corresponding protein factors during these alternative splicing events remain to be determined. Another type of alternative processing involves the inclusion or removal of various intronic sequences. Such is the case for the bovine growth hormone gene, in which the last intronic sequence may be retained in a fraction of mRNA and transported to the nucleus, allowing for production of a variant form of the hormone.24–27 Additionally, the use of alternative polyadenylation signals from a single transcript increases the diversity of its biological responses, as with the hormone calcitonin, which is produced in the thyroid gland, and calcitonin gene-related peptide, which is produced in the hypothalamus.2,22,25,28,29 Both hormones are the products of a single gene that undergo alternative processing and polyadenylation of its RNA transcript.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree