Summary of key facts
- •
The human microbiota is composed of bacteria, viruses, fungi, yeast, protozoa, and archaea. Because of its commensal relationship with the host, it is identified as a functioning organ system.
- •
The gut microbiota has been shown to have a critical role in immunotherapy outcomes.
- •
Colonies of the microbiota are found on the surfaces of the digestive tract, respiratory system, urogenital tracts, and skin.
- •
The human microbiota has more than 1000 different microbial species.
- •
An estimated 3.8 × 10 13 actual microorganisms can be found in the human gastrointestinal tract. More important, the varied microbiota of the gut express more than 3 million genes.
- •
Bacteroidetes and Firmicutes phyla account for an estimated 90% of the bacterial colonies in the gastrointestinal tract. Actinobacteria, Cyanobacteria, Fusobacteria, Proteobacteria, and Verrucomicrobia phyla constitute the remaining 10%.
- •
The initial microbiota of the neonate is acquired by vertical transmission from the mother and is thus accompanied by an inherited tolerogenic immunity to the colonizing microbiota.
- •
The early seeding and maturation of the newborn microbiota is critical to the development of a healthy, robust immune response system.
- •
The commensal relationship and integrity of the immune system goes on throughout life and is especially subject to any dysbiosis within the gut microbiome.
- •
Certain gut microbes and their metabolites have the ability to alter somatic cell DNA, resulting in cells with a malignant potential. An estimated 15% to 20% of human cancers have microbial and viral origins.
- •
Clinical studies have demonstrated that patients with lung cancer and kidney cancer having a poor response to treatment with PD-1 blockade had low levels of the gut bacterium Akkermansia muciniphila.
- •
Responders to anti-PD-1 blockade in this study had more abundant gut Bifidobacterium longum , Collinsella aerofaciens , and Enterococcus faecium than nonresponders.
- •
Clinical observations concerning the importance of the gut microbiome to achieving an effective ICI response have been supported by experiments in a number of ICI murine tumor models.
- •
Adoptive T-cell transfer anticancer immunotherapies, including CAR T cells, involve pretreatment lymphodepletion and the use of broad-spectrum antibiotics. Such additions to the therapeutic regimen are disruptive to the gut microbiome.
- •
In early studies, patients more likely to respond to anti-CD19 CAR T-cell therapy were found to have a relative abundance of Ruminococcus , Bacteroides , and Faecalibacterium pretreatment gut bacteria.
- •
Those who did not respond well were observed to have a greater abundance of Peptostreptococcaceae and Clostridiales.
- •
Fecal microbiota transplantation (FMT), accepted treatment for resistant C. difficile infection, is being applied in early clinical trials to generate checkpoint inhibitor responses in patients previously resistant to treatment.
- •
Clinical studies suggest that FMT can restore a patient’s ICI nonresponder taxa to that of a responder gut microbiome.
- •
Management of the gut microbiome appears critical to the efficacy of immunotherapy, and both clinical studies and animal models demonstrate the microbiome’s ability to directly influence the adoptive immune antitumor response.
- •
FMT holds significant promise as an effective and safe method to enhance immunotherapy responses and duration of responses and to treat irAEs that occur.
Introduction
I recall during my directorship of the National Cancer Institute (2006–2010) being appointed as one of several National Institutes of Health (NIH) institute directors to serve on an NIH external advisory committee charged with advising the NIH director regarding areas of science ready for prioritization. Consensus was quickly reached that the microbiome and its relationship to health and disease was primed for a significant research investment. , Discussion by those participating recognized that the organisms residing in the body such as bacteria, viruses, fungi, yeast, protozoa, and archaea, along with their genomes and metabolic products, actually can be considered an organ system in their own right. Though these varied organisms comprising the microbiota may be found virtually anywhere on the body, focus has been on sites such as skin and mucous membranes of the body’s external facing structures and the epithelial linings of the lungs, genitourinary tract, and gastrointestinal system.
As a result, in 2007 the NIH launched the Human Microbiome Project (HMP). The first phase of HMP (2007–2012) was focused on identifying and characterizing the human microbiota. , The second phase was launched in 2014 as the Integrative HMP (iHMP; http://ihmpdcc.org ) with the goal of generating integrated longitudinal data sets of microbiota biologic properties in association with disease conditions. An emphasis was placed on establishing the technologies of 16S rRNA gene profiling, whole metagenomic shotgun sequencing, whole genome sequencing, metatranscriptomics, metabolomics, and immunoproteomics as tools to study the human microbiota and how this complex system integrates as a functioning organ with the host. Current science in this field is greatly enabled by advances in computational biology, artificial intelligence, machine learning, and powerful computer systems capable of integrating large data sets.
The technologies originally developed to accomplish the Human Genome Project, as one can readily understand, were enabling to the iHMP. The iHMP was designed to be an interdisciplinary effort of disease-based or related projects. The goal of the iHMP has been to define the genetic and functional diversity of the organ specific microbiomes and their relationship to disease(s). The NMP and iNMP research projects demonstrated that each individual has their unique composition of microbiota strains. Though current studies in cancer have focused on bacteria, other microorganisms including the commensal viruses, fungi, and archaea cannot be neglected because they most certainly also have a role in cancer and immuno-oncology (for an in-depth review, see The Integrative HMP Research Netword Consortium, Voth and Khanna, and the NIH Human Microbiome Portfolio Analysis Team ).
Studies of the gut microbiome, for example, demonstrated the importance of maternal colonization and a period during newborn development with flora of low diversity, referred to as the period of “founder species.” The gut microbiome is seeded in utero and evolves early in life to a mature colonization. Composition of the gut microbiota is influenced by geography, ethnicity, diet, mode of delivery (vaginal vs cesarean section), illness, and use of antibiotics during the host’s early development. The intestinal microbiome is estimated to contain over 100 trillion microorganism cells that encode more than 3 million genes. The gut microbiota with its complex organization of microbes exists in symbiosis with the host and provides a remarkable array of protein products that play a critical role in host physiology.
During the past decade as a result of the NIH investments in the NMP, research has found increasing evidence for a strong relationship between the gut microbiome and cancer, including colon and rectum, esophageal, gastric, pancreatic, lung, melanoma, breast cancer, as well as hematologic malignancies. In this chapter, we will review what is known regarding the role the gut microbiome plays in the initiation and progression of cancer as well as its effect on cancer treatment especially immunotherapy.
The microbiome and immune system development
The bacterial component of the human microbiota is by far the most researched of the microbiota communities. These microbiota communities consist of bacteria, archaea, bacteriophages, viruses, fungi, and protozoa. The colonies of the microbiota can be found on the surfaces of digestive tract, respiratory system, urogenital tracts, and skin. , It is demonstrated that the host immune system, both innate and adaptive, evolved along with the colonization of the host microbiota to establish a healthy coexistence between the organisms of the microbiota and the responding capability of the host immune system. , This evolving relationship actually begins in utero and reaches a mature status somewhere between the ages of 3 and 5 years. This commensal relationship and the integrity of the immune system, however, are ongoing throughout life. The integrity and cellular makeup of the immune system is subject to alteration or impairment throughout life as a result of changes in the microbiota. Gut microbiota forces impacting the integrity of the immune system may be as simple as host dietary changes and as complex as may occur with the onset of certain chronic diseases and the administration of antibiotics , (for an in-depth review, see Maynard et al. ).
In the gastrointestinal (GI) tract, the Bacteroidetes and Firmicutes phyla account for an estimated 90% of the bacterial colonies. The remaining 10% is made up of colonies of the phyla Actinobacteria, Cyanobacteria, Fusobacteria, Proteobacteria, and Verrucomicrobia. The greatest bacterial density in the GI tract occurs in the colon. Research using advanced genomic techniques has demonstrated that the GI microbiome actively affects a number of host functions including metabolism, immunity, the development and progression of cancer, and the host responses to pharmacogenomic interventions. ,
The actual development of the immune system begins in utero, as does the growing relationship of the immune system with the developing fetal microbiota. Because the initial microbiota of the neonate is acquired by vertical transmission from the mother, there exists an inherited tolerogenic immunity to the colonizing microbiota. It has been demonstrated that transvaginal delivery is also an important source of seeding the newborn microbiome with the mother’s native and tolerated vaginal microbiota. Breast milk is also an important source of immune regulating factors involved in the early education and formation of the newborn immune system. Breast milk contains a number of immune stimulating factors such as actual live bacteria, protein metabolites, immunoglobulin (Ig) A antibodies, certain immune cytokines, and even cells of the immune system. Accumulating evidence has confirmed the critical role the early seeding and maturation of the newborn microbiota has to the postnatal imprinting and maturation of the immune system. In a sense, the constant priming of the host immune system by an intact microbiome is critical for an immune system ready to optimize the host’s ability to effect efficient antitumor therapies. Any effect on the normal development of the newborn microbiota can affect host immunity throughout life. , , ,
Conditions that occur in the host required for the host to eliminate an invading pathogen can become a risk to the host if these immune responses become chronic in nature. Such chronic inflammatory responses have the potential to cause dysbiosis within the microbiota and as a result alter host immunity. Host efforts to curb inflammation can be detrimental to the host’s antitumor response. The host’s efforts to curb inflammation can be thought of as being competitive to the host’s capacity to adequately mount a robust antitumor response during the early stages of tumor initiation (for an in-depth review, see Belkaid and Harrison ).
The microbiome and cancer initiation and progression
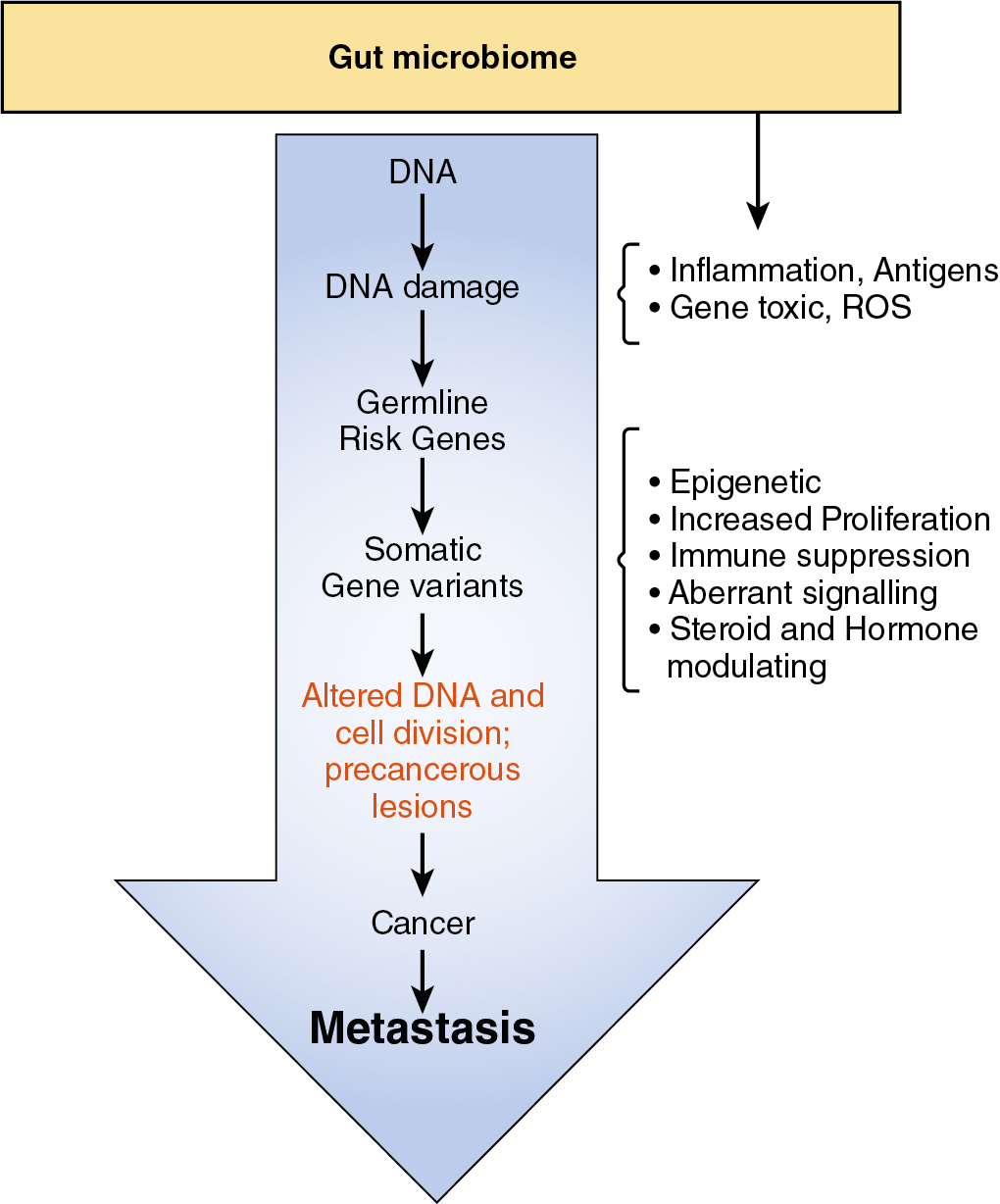
Over the past two decades, there has been increasing evidence that the intestinal microbiota has a critical relationship with the human peripheral immune system. As noted previously, more than 1000 different microbial species and an estimated 3.8 × 10 microorganisms can be found in the human gut, mostly in the lower colon. This varied microbiota expresses more than 3 million genes, the products of which have significant metabolic activity affecting normal host physiology and health. When there is a shift or dysbiosis in the gut microbiota composition causing disruption of the normal host microbiota homeostasis, the host is at risk for a broad array of altered physiology and even the onset of chronic diseases including cancer. , A dysbiosis within the GI tract may also significantly affect the immune system’s normal ability to eliminate cancer at its earliest stages of formation by suppressing innate immune surveillance and the adaptive antitumor immune response.
In addition to a dysbiosis that may impair normal immune surveillance and an anticancer immune response, there is accumulating evidence that certain gut microbes and their metabolites have the ability to alter DNA in somatic cells of the host. Alterations of somatic cell DNA may interfere with the host’s genes responsible for controlling the cell cycle, causing increased cell proliferation, and also generate metabolites that disrupt normal cell death programs. Alterations in the GI tract microbiota and the resultant induced events in DNA coding and in gene transcription increase the risk for the development of somatic cells with malignant potential.
Perhaps the best-known pathogens directly involved in the inflammation–cancer pathway are Helicobacter pylori in gastric cancer, human papillomavirus in cervical and head and neck cancer, and hepatitis B and C viruses in hepatocellular cancers ( Table 11.1 ). , , ,
| Class I Microbes | |
|---|---|
| Carcinogenic Microbe | Site of Cancer |
| Helicobacter pylori | Stomach |
| Liver |
| Human papilloma virus | Cervix, vagina, vulva, anus, penis, oropharynx |
| Epstein-Barr virus | Nasopharynx, non-Hodgkin lymphoma, Hodgkin lymphoma |
| Kaposi sarcoma–associated herpes virus |
|
| Human T-cell lymphotropic virus type I | Adult T-cell lymphoma |
| Bladder |
In addition, certain pathogens can promote inflammation and bacterial metabolites that directly trigger tumor initiation. An estimated 15% to 20% of human cancers have been connected either directly or indirectly to having microbial origins. , Enterococcus faecalis , for example, produces reactive oxygen species responsible for the formation of a DNA-damaging compound. Important to the relationship between the microbiome and the antitumor immune response is the observation that bacterial microbes originating in the gut can be found in both the tumor and the tumor microenvironment, where they secret factors that promote tumor cell growth and suppress antitumor immunity ( Table 11.2 ). , ,
| Bacterial Metabolites | Potential Cancer-Causing Action |
|---|---|
| Sulfate-reducing bacteria (H 2 S) | H 2 S has cytotoxic and genotoxic actions. |
| Nitrate reduction to nitrite; N-nitroso compounds | Multiple gut bacteria involved, can form DNA adducts |
| Polyamine production such as ornithine | Multiple gut bacteria are capable; associated with increased inflammatory tumor microenvironment |
| Secondary bile acids, deoxycholic acid, lithocholic acid | Multiple Clostridium sp., tumor-promoting activity |
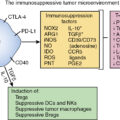
Alterations in the gut microbiota have been shown to promote inflammation in the intestinal tract and epithelial cell proliferation through the activation of the interleukin (IL)-6 signaling pathway. Epithelial inflammation results in microbial-generated products such as lipopolysaccharides (LPS) causing immune cell proliferation and migration along with an upregulation of Toll-like receptors (TLRs). Bacterial products produced in excess as a result of gut dysbiosis and inflammation can activate nuclear factor kappa B (NF)-κB, c-Jun N-terminal Kinase (c-Jun/JNK), and Janus Kinase/signal transducer and activator of transcription (JAK/STAT3) pathways, all of which have well-demonstrated actions causing abnormal cell proliferation and suppression of the antitumor immune response ( Fig. 11.1 ).
The microbiome and cancer immunotherapy
Spontaneously arising cancers in humans are well recognized for the development of their capacity to avoid immune surveillance and to withstand tumor cell destruction by the adaptive immune response. Immunotherapy first became an option for treatment of malignant diseases with the introduction of monoclonal antibodies and the use of stem cell transplantation. , The ability to reverse tumor cell resistance to the host antitumor immune response has been demonstrated with the blocking of immune checkpoints responsible for down regulation of the antitumor T-cell response (ICI). ICI immunotherapy involves using monoclonal antibodies against the cytotoxic T lymphocyte–associated protein 4 (CTLA-4) and against the programmed cell death protein 1 (PD-1) or its programmed cell death ligand 1 (PD-1L) immune checkpoints. These monoclonal antibody inhibitors have had rather impressive responses and represent the first real breakthrough in immunotherapy. As a result, ICI has become the standard of therapy for a number of solid tumors and hematologic malignancies.
In addition to the revolutionary success with ICI, a variety of cell-based therapies including adaptive T-cell therapy (ATC), in vitro expansion of tumor-infiltrating lymphocytes (TILs), T-cell receptor–engineered T cells (TCR), and chimeric antigen receptor gene–transduced T cells (CAR T cells) have demonstrated promising cell-based antitumor efficacy and are currently the subject of intense investigation. ,
The intestinal microbiome and ICI immunotherapy
Two observations have suggested that the response to ICI may be more complex than the simple antibody blocking of the cellular checkpoint. The first observation suggested that patients with cancer with a high body mass index (BMI) have a significantly higher ICI response rate than that observed for patients with a normal or low BMI. , The second observation found that patients with cancer who had received courses of antibiotics immediately before or early in their ICI treatment had much lower ICI response rates than those without an antibiotic use history. This accumulating evidence therefore began to support the conclusion that ICI nonresponders may have an imbalance in gut flora composition that correlates with impaired immune cell activity (for an in-depth review, see Frankel et al. ).
Routy and colleagues reported their studies of fecal samples from patients with lung cancer and kidney cancer being treated with checkpoint inhibitors. They discovered that those patients having a poor response to PD-1 blockade had low levels of the bacterium Akkermansia muciniphila . In 2018, Matson and colleagues reported similar studies of patients with progressive metastatic melanoma undergoing anti-PD-1 blockade. In these studies, they analyzed baseline stool samples before ICI therapy using integrated analysis of 16S ribosomal RNA gene sequencing, metagenomic shotgun sequencing, and quantitative polymerase chain reaction (qPCR) of selected bacteria. They found that responders to ICI had more abundant bacterial species of Bifidobacterium longum , Collinsella aerofaciens , and Enterococcus faecium than nonresponders. They also used germ-free mice whose GI tracts were reconstituted with fecal material obtained from ICI-responding patients with melanoma. The mice demonstrated improved tumor ICI response and tumor control and greater T-cell immune responses.
In another report examining the oral and gut microbiome of patients with metastatic melanoma being treated with anti-PD-1 immunotherapy, analysis of 30 ICI responders and 13 nonresponders found that responders had significantly greater alpha diversity ( p < 0.01) and a relative abundance of bacteria of the Ruminococcaceae family ( p < 0.01). In the nonresponders the Bacteroidales order was found to be enriched. Metagenomic WGS also showed enrichment of Faecalibacterium species in responders and nonresponders were enriched for Bacteroides thetaiotaomicron , Escherichia coli , and Aaerotruncus colihominis . The investigators supplemented these initial clinical studies by using fecal microbiota transplantation (FMT) from patients to germ-free mice. They concluded that nonresponders had an unfavorable gut microbiome with low diversity and an abundance of Bacteroidales leading to an impaired systemic antitumor immune response. They showed that the impaired immune response resulted from impaired lymphoid and myeloid cell tumor infiltration and a suppressed capacity for antigen presentation. There have been a number of studies directed at characterizing the ICI responder gut taxa and how these species relate to the antitumor immune response ( Table 11.3 ).
| ICI Immunotherapy | Involved Microbes | Mechanisms of Effect on Outcome |
|---|---|---|
| Anti-CTLA-4 mAbs | Bacteroides fragilis, Bacteroides thetaiotaomicron, and Burkholderia cepacia | Stimulation of CD1b + DCs increased IL-12-dependent Th1 antitumor response. |
| Bifidobacterium | Enhanced suppressive activity of Tregs decreasing immunopathology caused by ICI. , | |
| Bifidobacterium pseudolongum and Akkermansia muciniphila |
| |
| Anti-PD-1 and PD-L1 | Bifidobacterium breve and Bifidobacterium longum |
|
| Akkermansia muciniphila | Increasing TCM, CD4/Foxp3 ratio in tumor and TME, increases IL-12 and IFN-γ production, resulting in synergistic effect with PD-1 blockade. , | |
| Lactobacillus rhamnosus GG | Activates DCs via cGAS-STING-TBK1-IRF7-IFN-β cascade to enhance CD8 + T-cell activity against cancer cells. | |
| Bifidobacterium breve | Molecular mimicry between SVY antigen of B. breve and SIY neoantigen of mouse melanoma stimulated cross-reactive T-cell response against melanoma cancer cells. | |
| Bacteriophage-infecting Enterococcus hirae | TMP of bacteriophage stimulated memory CD8 + T-cell cross-reactivity with cancer antigen PSMB4 protein. |
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree