Summary of key facts
- •
The natural history of a human malignancy and its relationship to the host immune system has been described as comprising the “three Es”: elimination, equilibrium, and escape.
- •
In order to progress, the tumor needs to escape immune control by the cell destructive capacities of the CD8 + T cells of the adaptive immune system and of the NK cells of the innate immune system.
- •
While attempting to eradicate the tumor, the immune system undergoes what has been described as a process of immunoediting to alter the immune system’s recognition of the tumor as nonself, resulting in the tumor escaping immune destruction.
- •
Despite initial immune recognition of the tumor, as the tumor progresses, clones of tumor cells with lower capacity for antigen recognition and/or greater resistance to immune cell destruction are preferentially selected for tumor progression (immunoediting).
- •
The many diseases called cancer are characterized by their genetic instability and, as such, are prone to mutations and structural genetic variants occurring in the genes coding for proteins responsible for performing the many normal processes of the cell, including the complex mechanisms of transcription and translation. A tumor cell may have more than 1000 genetic variants, confirming the complexity of cancer as a disease.
- •
Tumor cells can avoid immune recognition and destruction in multiple ways:
- •
a silencing of intrinsic tumor cell processes to generate and to select for MHC presentation of strong antigenic peptides (TSAs and TAAs).
- •
a silencing of or mutations in the HLA genes of MHC class I and MHC class II molecules.
- •
by co-opting immune checkpoint processes to enable immune suppression.
- •
by recruiting inflammatory cells to the tumor and TME with immunosuppressive capacity.
- •
by molecular alterations to intrinsic T-cell signaling pathways.
- •
- •
Genetic alterations in the control mechanisms for mRNA translation can be a cause of tumor antigen loss despite normal MHC antigen-presenting molecules.
- •
MHC class II antigen presentation requires the classic antigen-presenting cells, especially dendritic cells, to first take up tumor antigenic peptides released from tumor cells and internally process these for loading on the MHC class II molecule. The multiple steps required in this process provide ample opportunity for a failure to generate the stimulatory pMHC class II molecule for TCR presentation.
- •
Tumor-derived antigenic peptides can undergo self-tolerance within the thymus. This process appears to be selective for tumor antigenic peptides that survive tapasin editing for MHC class I presentation and DM editing for MHC class II presentation.
- •
Tumor cell activation of the WNT/β-catenin pathway is an example of signaling pathway interference with T-cell infiltration of the tumor and of the TME.
- •
The TME and tumor-associated chronic inflammation are closely involved in all stages of cancer development and progression.
- •
The presence of chronic inflammation in the TME is the driving source of immune suppression and the facilitator of tumor metastasis. TME inflammation is a key factor in therapeutic efficacy.
Introduction
We have come to understand the many diseases we call cancer by their genetic and epigenetic instability and the relationship the growing tumor has with its surrounding cellular microenvironment and with the host’s extensive immune system. Genetic instability in the tumor leads to the development of tumor-specific neoantigenic peptides and, on occasion, ectopic overexpression of genes not normally expressed in the tissue in which the tumor has developed. These non-self-antigenic peptides have an opportunity for presentation by major histocompatibility (MHC) class I molecules expressed on the tumor cells and for cross-presentation by MHC class II molecules expressed by classic antigen-presenting cells. The peptide-loaded MHC molecules present the antigenic epitopes to the T-cell population of the adaptive immune system and to natural killer (NK) cells of the innate immune system. The level of antitumor response by both innate and adaptive compartments of the immune system depends on numerous interactions between the tumor cells and various immune cells and with inflammatory cells of the tumor microenvironment (TME). Antitumor immunity occurs at multiple levels within the host, both locally at the tumor site and peripherally in the immune system. The patient’s antitumor response must react to a changing tumor burden and to therapeutic interventions requiring constant efforts of response renewal.
The intimate relationship between the tumor and the immune system occurs over the entire course of disease, from initiation through the progressive stages of growth, local invasion, and metastasis to distant sites. For tumors to survive and progress along this path requires that the tumor develop mechanisms that actively inhibit antitumor cell immune responses. , Much like the complexities of the antitumor immune response, there are multiple, equally complex mechanisms that function to enable the tumor cells to resist immune destruction. , Though much has been learned regarding the development of immune resistance to avoid immune tumor cell destruction, developing effective immune therapies that overcome this resistance remains a tremendous challenge. This chapter will address the intimate relationship between the tumor as it progresses and interacts with the patient’s immune system with a focus on the mechanisms that enable the tumor to evade immune destruction.
The presence of tumor immune suppression
The concept of the three “Es” of cancer—elimination, equilibrium, and escape—was first introduced in 2004 as a way of describing the relationship between the initiation and development of cancer and the host’s immune system. In the elimination phase, potentially malignant cells arising during the early initiation stage of cancer development are detected as nonself and eliminated as a result of their MHC class I presentation of neoantigens. They are recognized as foreign by the NK cells of the host’s innate immune system and by cytotoxic T lymphocytes (CTLs) of the adaptive immune response. This stage has been equated with the historic concept of immune surveillance. If the tumor cells escape elimination, they may enter the second stage, termed the equilibrium phase . In the equilibrium phase, surviving tumor cells, not eliminated by the immune system, are instead held in check by the immune system without being destroyed but without progression.
Most cancer clinicians recognize a second clinical component of this equilibrium phase. For example, it is not uncommon for aggressively treated newly diagnosed cancer to enter a period of complete response (CR) only to recur many years later. Clearly, these CR patients have undetected micrometastases after treatment and remain in a state of equilibrium or at least controlled slow progression. Clinically, we refer to this phase of cancer as disease-free survival (DFS) or in some cases stable disease (SD). During this phase of DFS or SD, the relationship between the tumor and the host’s immune system is one of controlled equilibrium. Finally, should the tumor progress to the third escape stage, its capacity to be immunogenic has been significantly altered and on balance it is no longer completely susceptible to immune cell destruction by the host.
As the tumor progresses through these three stages, it is said to undergo immunoediting. Immunoediting has been described as the complex process(s) by which the host immune system can either inhibit tumor growth or be suppressed/edited in ways that promote tumor progression. There are multiple targets and intracellular pathways that provide mechanisms by which tumor cells can escape both the innate and adaptive immune systems. These escape mechanisms will be described individually but almost certainly occur in a combinatory fashion for successful tumor escape ( Box 8.1 ).
Genetic downregulation or loss of MHC class I and II molecule expression.
Downregulation or genetic alterations in mechanisms controlling immunogenic antigenic peptide creation and MHC antigenic peptide loading.
Upregulation of immune checkpoint ligands inhibitory to CTLA-4 and PD-1 receptors expressed on NK cells, CD8 + CTLs, and CD4 + T cells.
Antigen loss: silencing of or mutations altering tumor antigenic epitope translation.
Tumor-derived immune suppression: membrane bound and secreted.
TME chronic inflammation suppression of antitumor immune response.
Downregulation of or actual loss of MHC class I and II expression
As presented in Chapter 5 , the MHC class I molecules are the mechanism by which nonself tumor neoantigenic peptides are presented on the surface of tumor cells to the CD8 + T cells of the adaptive immune system. Though MHC class I molecules are essential for the presentation of foreign antigens such as infectious viral peptides or cancer antigens, conditions in which the expression of the MHC molecules is structurally defective or even absent are not uncommon in the genetically altered cancer cell. Importantly, defective or absent tumor cell MHC molecules does not interfere with tumor cell survival. It is recognized that tumor cell surface MHC class I neoantigen presentation is generally not a very efficient method of tumor antigen T-cell stimulation. In fact, cross-antigen presentation by MHC class II molecules expressed on antigen-presenting cells (APCs) is generally the more effective mechanism for stimulating the CD8 + T cells and other T cells of the immune system. Tumor antigen cross-presentation requires the antigenic peptides produced in the tumor cell to be released and taken up by APCs such as the dendritic cells (DCs) and presented to T cells via the MHC class II molecules rather than MHC class I.
Nevertheless, any impairment in tumor cells to the normal expression of genes involved in the processing of potential tumor antigenic proteins and in their presentation to the T-cell receptor (TCR) of the CD8 + T cells will have a significant negative effect on the host’s antitumor response. For example, let us begin with the dysregulation of the MHC chromosome 6p region’s human leukocyte antigen (HLA) genes. The class I region of the polymorphic HLA locus includes, among others, the highly polymorphic HLA-A , HLA-B , and HLA-C genes ( Fig. 8.1 ). These genes generate the 45 kDa classic α-chain (362–366 amino acids) of the class I molecule. The MHC class I β 2 microglobulin chain of 12 kDa is coded for by a gene on chromosome 15, a site different from the HLA region located on chromosome 6p21.3. Two of the three class I α domains, α 1 and α 2 , create the MHC domain containing the neoantigen peptide-binding cleft. The class II MHC molecule has a similar overall physical structure but is the product of two HLA genes containing one α 1 and one β 1 domain that fold to generate the groove that holds the antigenic peptide (for an in-depth review, see Monos and Winchester ).
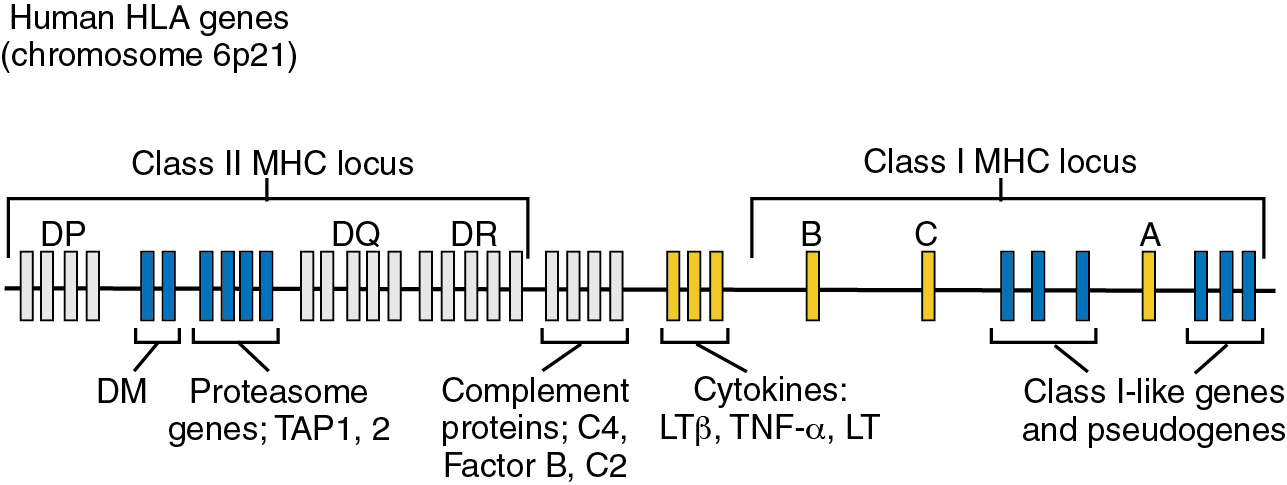
It is easy to understand that within the genomically unstable tumor cell there is ample opportunity for somatic mutations to occur in the HLA genes, resulting in aberrant expression or actual silencing of HLA class I and II molecules. An example of HLA-I gene dysregulation and dysregulation of genes involved in antigen processing and presentation can be found in studies at the University of Cologne reported by Thelen and colleagues. They determined the expression level of 24 genes known to be involved in antigen presentation. The genes were selected based on their known downregulation in at least a portion of patients for one or more of the tumor types selected.7, They found that the mean expression levels for 7 of 24 genes were increased greater than twofold in the tumor microenvironment (TME) of tumor samples compared with normal tissues. They selected a twofold change as a cutoff in order to determine evidence for impaired gene expression. They found TME downregulation to occur in at least one patient of the following genes β2M , HLA-A , HLA-B , HLA-C , ERAP1 , ERAP2 , PDIA2 , NLRC5 , UBB , UBC , and LMP10 . This downregulation was reported in >10% of analyzed tumor samples and occurred in more than 3 (>12.5%) of the included genes in 45 of 142 patients (32%). No alterations in expression level were found in 23% of patients. They noted that expression patterns of the 24 genes across tumor types were quite heterogeneous. Though this represents an early effort to show HLA-I gene dysregulation as an important aspect of immunoediting during cancer progression, it clearly emphasizes both the complexity and the critical relationship of HLA expression to immunotherapy.
The loss of both copies of MHC class I heavy chain genes or of β2M will, of course, eliminate the expression of a major component of the antigen presentation molecule and has been observed in cancers such as melanoma with the loss of HLA-A2 expression. Many cancers have also been found to have a loss of only one copy of the heavy chain or of only β2M (heterozygous). , MHC class I heavy chains are codominantly expressed but the genetic alteration at the level of the heavy chain gene may be heterozygous. Montesion and colleagues surveyed 59 different cancer types and observed that in 17% there was a significant loss of MHC I expression secondary to a loss of heterozygosity , (for an in-depth review, see Dhatchinamoorthy et al. ).
There are a few facts to keep in mind regarding the function of MHC class I molecules. First, as part of their normal functions, all cells generate essential proteins and, as a result, present short peptides derived from those proteins on their cell surface as part of the peptide–MHC (pMHC) complex. When the peptide is nonself as in cancer, it is recognized as such by the immune system—innate or adaptive—for the cell to be destroyed. In all cells, the expression of the MHC complex on the cell surface is governed by interferons, especially interferon gamma (INF-γ) and the IFN signaling pathway. Stimulation with INF-γ increases the expression of MHC class I and therefore of detectable MHC class I molecules on the surface of tumor cells. The loss of MHC in cancer or the suppression of MHC expression correlates significantly with the progression of the cancer, indicating the significance of this mechanism in tumor resistance and poor prognosis ( Fig. 8.2 ).
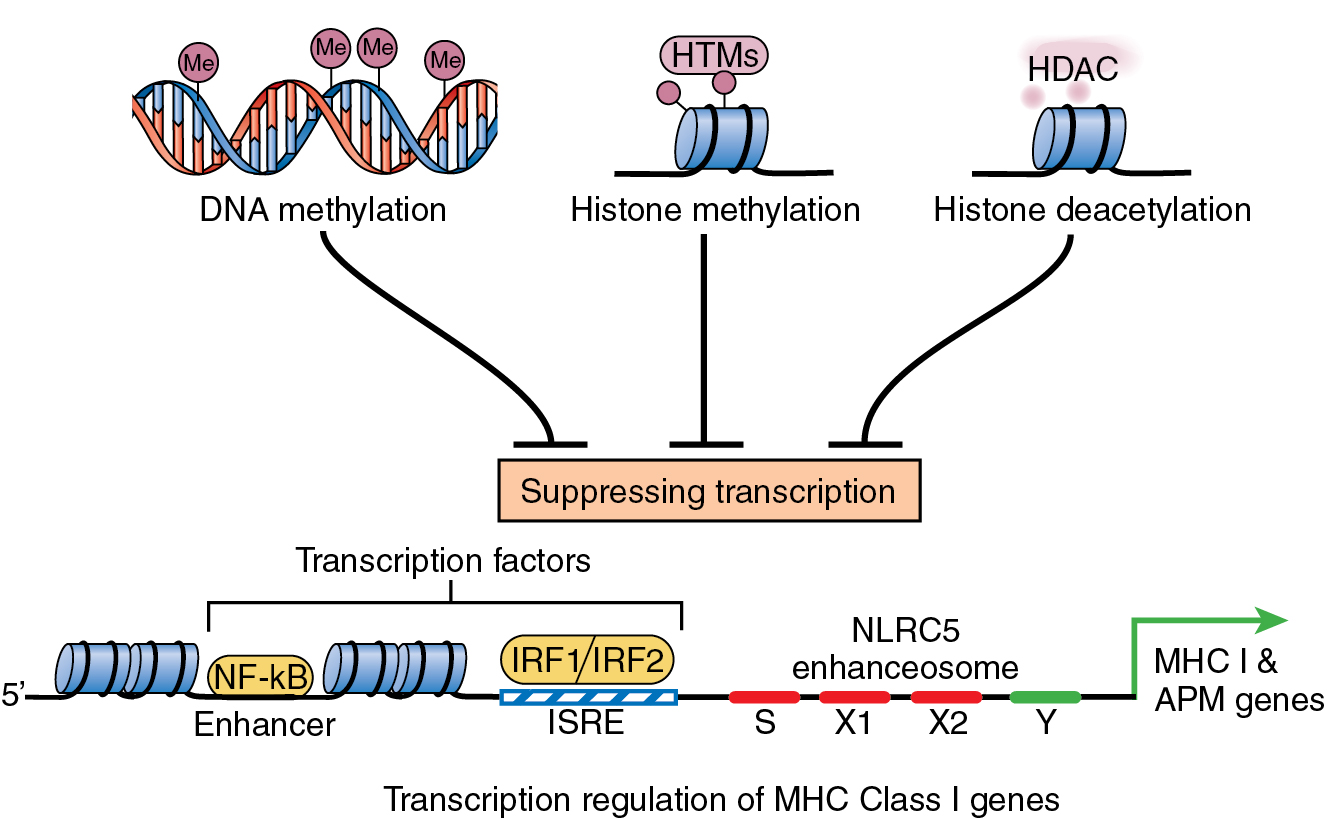
MHC low expression metastases are also known to respond poorly to immunotherapy. Many genes coding for proteins intimately involved as factors in the ubiquitin–proteasome production of the neoantigenic peptides can also be subject to genetic alteration in the tumor cell. Such genetic alterations in the proteins comprising the processing machinery of the proteasome can alter the neoantigenic peptide, causing a loss of its antigenic epitope or simply a loss of the peptide altogether. Such alteration can also occur as the potential antigenic peptides leave the proteasome and enter the endoplasmic reticulum where they are loaded on the MHC class I molecules (see also Chapter 5 , Fig. 5.2 ).
Many of the genes involved in the preparation of immunogenic peptides have similar transcription and translation governance, and many of the genes for these processing factors are actually chromosomally located in association with the HLA heavy chain genes on chromosome 6p. Thus genetic alterations affecting these genes may manifest in low expression of MHC molecules. Studies show that within the tumor itself there exists significant tumor cell heterogeneity in terms of MHC expression and therefore the robustness of T-cell antigen presentation. This heterogeneity in levels of pMHC cell expression is also found in relationship to the various metastases of a given tumor both as to the metastatic site (lung, liver, brain, etc.) and within the actual individual metastases. Low or absence of detectable MHC class I has been reported to varying degrees in patients for many of the most common human malignancies including breast, prostate, colorectal, melanoma, head and neck, and non-small cell lung cancer (NSCLC), for example. It also appears that the level of expression may vary over the course of disease and in association with therapy. Finally, a loss of MHC class I has been reported to be associated with resistance to anti-programmed cell death protein-1 (PD-1) immunotherapy. ,
It is easy to understand how in a genomically unstable tumor cell there exists ample opportunity (see Chapter 5 ) for genetic variants to cause defects that prevent optimal MHC molecule formation and optimal tumor antigen generation suitable for pMHC class I presentation. For example, any loss of the key molecules such as transporter for antigen processing (TAP), tapasin, or endoplasmic reticulum aminopeptidases (ERAP1 and ERAP2) will cause a loss of pMHC expression and presentation on the cell surface. Tumor cells that sustain homozygous deletion or frameshift variants of the HLA region of chromosome 6p will result in low expression or complete loss of MHC class I antigenic peptide presentation. Some alterations are unique to a specific cancer, whereas others may be found in numerous patients and in a variety of cancer types ( Box 8.2 ).
Mutations or deletions of HLA structural genes.
Variants in genes or loss of genes involved in the tumor antigen MHC presentation pathway.
Variants affecting the transcription of genes critical to the pathway.
Epigenetic silencing of pathway gene regulatory elements.
Variants affecting mRNA fidelity and/or stability in generating pathway elements, pre and posttranscription.
A number of clinical correlations with low or absent MHC class I are important to note. For example, the presence of low MHC class I expression in breast cancer is associated with tumors that contain fewer tumor-infiltrating lymphocytes (TILs) than the tumors demonstrating a high degree of MHC class I expression. Second, though the low expression of pMHC molecules or even a total absence prevents activation of the T cells and triggering of adaptive immune response against the tumor, it does not have a negative effect on cancer cell viability or tumor cell proliferation. The low expression or loss of MHC, as expected, does strongly indicate a worse cancer prognosis. Third, and perhaps even more important, is the association of low or absent MHC class I expression with development of resistance to anti-PD-1 and anti-PD-L1 immunotherapy. A similar negative correlation is associated with MHC class I loss in other epithelial tumors such as melanoma, cancers of the colon and rectum, and head and neck cancer , (for an in-depth review, see Gettinger et al. ).
MHC class I molecules are more densely presented on the cells of the hematologic lineage. MHC class II molecules, in contrast, are predominantly found on the surface of antigen-presenting cells such as dendritic cells (DCs), B cells, and classic macrophages. Their major role is the presentation of antigenic peptides such as from viral infections and cancer cells produced in other cells from which these peptides are released. The released antigenic peptides are internalized by the APCs for further processing and for loading onto the MHC class II molecules for surface presentation. The antigenic peptide–MHC class II molecule is primarily interactive with the CD4 + T cells. CD4 + T cells function to support and enhance the CD8 + CTL of the adaptive immune response and to generate effective memory T cells.
In addition to a direct interaction of the antigenic peptide bearing tumor-specific MHC class II molecule (tsMHC class II) with the TCR of CD4 + T cells, tsMHC class II molecules have been implicated in the stimulation directly or indirectly of other T-cell subsets. Secretion of several known activating cytokines, such as IFN-γ, can be triggered by tsMHC II interaction with CD4 + T helper (Th) cells. On the other hand, the stimulation of CD4 + T cells may result in activation of the subclass of regulatory T cells (Tregs), which function to suppress the immune response favoring cancer progression.
Some tumor cells during the cancer’s progression have been shown by immunofluorescence to actually present both MHC class II and MHC class I molecules. Tumor-specific MHC class II expression may increase the immune response to the tumor. In fact, this finding has been associated with improved prognosis in response to immunotherapy. The binding groove of the MHC class II molecules can bind longer peptide chains than can be held in the groove of the MHC class I molecule and also accept peptide chains having more branched side chains that are more immunostimulatory when interacting with the TCR.
The MHC class II molecule is heterodimeric in contrast to MHC class I, and studies have demonstrated that expression of the class I and class II molecules is independently regulated in cancer. This independent regulation may in part account for observed variations in immunotherapy responses. For example, MHC class I molecular assembly and expression are not normally controlled except in T cells. Its expression, however, can be induced and enhanced by IFN-γ and by activation of the NF-κB signaling pathway. The expression of MHC class II, in contrast, is tightly controlled by the “master transcription control factor,” a protein (class II major histocompatibility complex transactivator) encoded by the CIITA gene located on 6p.
There are a number of intracellular alterations in signaling pathways that can contribute to the suppression of MHC class II expression. For example, JAK/STAT cell signaling is necessary for upregulation of MHC class II molecule expression and, as such, mutations occurring in JAK result in suppressed IFN signaling and suppressed expression of MHC class II molecules. Also, with activation of RAS/MAPK signaling in breast cancer, there is suppression of MHC class II expression. Clearly, from clinical studies the extent to which MHC class II expression is suppressed is a strong biomarker for a poor response to immunotherapy. ,
In summary, increasing evidence from clinical studies of solid tumors suggests the following: (1) the presence of a significant increase in tsMHC class II expression is associated with an improved disease-free survival (DFS) and overall survival (OS) perhaps as a result of increased antigenic recognition by the adaptive immune system, (2) significant surface expression of tsMHC class II is required for CD4 + T-cell (including T-cell subsets Th1 and Treg) activation, and (3) the increased presence of tsMHC class II correlates with improved response to anti-PD-1 and anti-PD-L1 immunotherapy. The apparent significant role that the neoantigen-loaded MHC class II molecule plays in the immune control of the tumor’s progression highlights why it is extremely important when anticipating the use of immunotherapy to determine whether the tumor is showing a low or possible absence of tsMHC class II expression or an intermediate or high level of expression, as well as upon which cell types the expression is occurring. This information is increasingly predictive of likely response.
Immune checkpoints and checkpoint ligands
Tumors, as noted, not only escape recognition and destruction by the host’s immune system but can actively block normal T cell–directed antitumor activity. One such method is by modulating immune inhibitory signals termed immune checkpoints . The immune checkpoints and their biologic pathways represent one of the most exciting anticancer therapeutic opportunities since their introduction in 2011 and, as such, one of the most intensely studied. In normal health, these immune checkpoints operate to prevent immunity to self (autoimmune diseases) and to self-proteins when the host is responding to infectious pathogens.
Cytotoxic T lymphocyte–associated antigen 4 (CTLA-4) was the first immune checkpoint receptor to be studied clinically. CTLA-4 functions primarily to depress the early T-cell response to tumor neoantigens by countering the TCR costimulatory receptor CD28. , Antigen recognition by the TCRs of T cells and the subsequent activation of CD8 + CTLs increases the expression of the CTLA-4 receptor on the T cells. TCR activation also suppresses the numbers of CD4 + T helper cells and their activity, along with an increase in numbers of CD4 + Tregs, which have an immunosuppressive effect. Taken together, these multiple actions have a suppressive effect on the antitumor immune response and favor tumor progression.
Another important checkpoint inhibitor is the programmed cell death protein 1 (PD-1). PD-1 receptor protein is expressed on T cells with the function of limiting T-cell activity in peripheral tissues at the time of pathogenic inflammation and of cancer initiation and development. In cancer, the ligand to PD-1 (PD-L1) is expressed in tumor cells and other cells of the TME. A high or enhanced expression of the ligand PD-L1 by the tumor causes increased stimulation of the T-cell PD-1 receptor, resulting in suppression of these T cells and their functions; in doing so, it promotes tumor growth. PD-1 activation generates suppression of two important antitumor activities. It triggers the induction of apoptosis to antigen-specific T cells and blocks the apoptosis of Tregs. PD-1 is a member of the immunoglobulin gene superfamily and is expressed as a transmembrane protein with a C-end cytoplasmic tail and an N terminus IgV-like domain. PD-1 is found to be highly expressed on TCR-activated tumor-specific T lymphocytes and is also found expressed to some extent on NK cells, B cells, dendritic cells, macrophages, and monocytes.
PD-1, in conditions where the immune T cells are challenged with a pathogen and inflammation, functions in the antigen-reactive T cell to prevent ineffective or harmful immune responses to self-tissues, helping to maintain a condition of immune self-tolerance. When expressed on immune cells that are being activated by cancer antigens, PD-1 has a very different role. When faced with cancer, PD-1 acts to interfere with the antitumor response by downregulating the T-cell responses, causing a decrease in T-cell proliferation and clonal expansion, a decrease in secretion of cytokines, and an increase in T-cell demise ( Fig. 8.3 ). Other described checkpoints include YIM-3, LAG-3, and VISTA. Targeting the immune checkpoints with blocking humanized monoclonal antibodies has emerged as a strategy to generate beneficial antitumor immune responses (for an in-depth review, see Han et al. ).
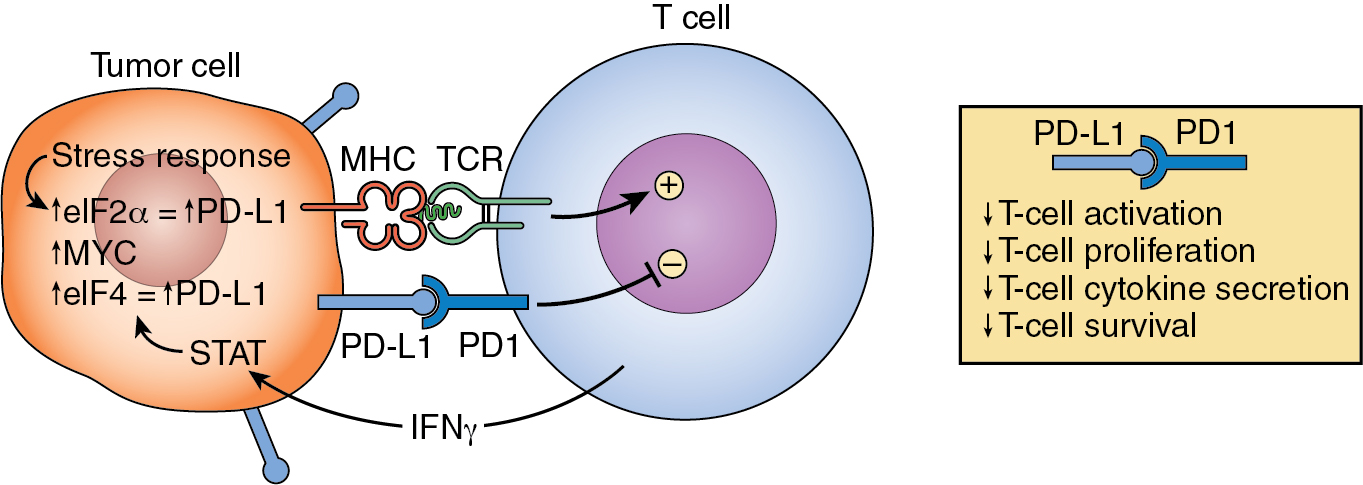
The ligand that binds to the PD-1 receptor, PD-L1, is expressed by cancer cells and, as such, plays the role of enabling the tumor cells to escape immune attack. PD-L1 is a small (33 kDa) B7 transmembrane glycoprotein with extracellular domains of Ig- and IgC. PD-L1 is also found to be expressed by DCs, macrophages, and B cells, as well as some epithelial cells, during inflammation. IFN-γ is known to increase the expression of PD-L1 in most cancers, and the pathway involved in PD-L1 expression also appears involved in several additional protumor stimulatory actions such as tumor cell proliferation, epithelial-to-mesenchymal transition, and expansion of the tumor stem cell population in some tumors. A presentation of all of the intrinsic cell signaling pathways involved in interacting to provide influence/control over the PD-L1/PD-1 axis in cancer is beyond the scope of this chapter but it has been well reviewed by others. Of increasing interest is how the genetic variants that define and alter the tumor microenvironment (TME) over time also manifest themselves in the up- and downregulation of various microRNAs that directly affect these signaling pathways. Undoubtedly, this will be a critical map to develop to optimally guide the integration of future immunotherapies.
Antigen loss variants—loss of mRNA translation
In Chapter 5 of this text, a good deal of effort was devoted to providing a detailed overview of the intracellular mechanisms involved in the generation and characterization of tumor-specific antigens (TSAs) and tumor-associated antigens (TAAs). As repeatedly noted in Chapter 5 and in this chapter, the genetically unstable cancer cell provides ample opportunity for the development of non-self-peptides containing antigenic epitopes of varying degrees of antigenicity when presented to CD8 + T cells. As presented, there is a complex multistep process involving many cytoplasmic proteins and enzymes required to fashion an appropriately sized antigenic peptide within the immunoproteasome and transporter molecules required to shuttle the peptide to and through the endoplasmic reticulum and to chaperone its loading into the MHC class I molecule’s groove for transport to the cell surface.
Mutations and structural variants, aspects of immunoediting, have the potential of affecting any of these factors directly or indirectly via variants that disrupt other critical signaling pathways. Any of these genetic alterations may affect this process, resulting in (1) selection of a short peptide with a weakly immunogenic epitope and (2) the generation or selection of an antigenic peptide that is not well sized or structured for optimal stable positioning in the MHC groove, interfering with optimal TCR recognition and T-cell immune activation. All throughout the process of tumor-specific neoantigen creation there are opportunities for mutations and structural variants to occur, resulting in a loss of optimal antigen presentation and a weakening of the tumor’s immunogenicity that facilitates the escape of the growing tumor from immune control. , Tumor antigen loss includes any of the possible alterations described that lead to a decrease or absence of the expression of any of the components required (1) to create and select an optimally antigenic short TSA peptide in the immunoproteasome, (2) for the formation of a competent MHC class I molecule for antigen presentation, and/or (3) for a loss of costimulatory molecules involved in T-cell activation as well as ligands for NK cell–activating receptors.
Studies have uncovered yet an additional cause of tumor-specific antigen loss. In these experiments, it has been demonstrated that tumor antigen loss may result from a unique posttranscriptional process by which tumor cells lose antigen expression by suppressing the translation of the tumor antigen’s mRNAs. There is increasing indication that RNA-binding proteins and microRNAs are involved in the active suppression of the translation of tumor cell TSAs. Evidence to support this mechanism of antigen loss can be found in a series of experiments reported recently (2019) by Han and colleagues using a CT26/HER2 murine protective tumor model to induce CTL and HER2 immunity. These experiments demonstrated that CT26/HER2 tumor cells could develop resistance to primed antitumor CTL through the loss of antigen expression while continuing to express MHC class I molecules. The authors concluded from their experiments that the CT26/HER2-A2 tumor cells lost antigen expression and tsMHC class I presentation secondary to a defect occurring at the posttranscriptional level. They demonstrated that the CT26/HER-A2 tumor cells expressed HER2 mRNAs but not HER2 antigenic proteins.
This observation of a tumor cell defect in translation should not be surprising, but certainly it requires a careful examination of the mechanisms involved and the extent of specificity involved in targeting tumor-generated neoantigens. The most critical and highly regulated step in mRNA translation is the initiation of translation by the eukaryotic initiation factors (eIFs). These initiation factors have a critical role to play in controlling both the specificity of mRNA translation and the rate of translation of a given mRNA. Thus, the initial step in translation involves the assembly of the appropriate protein factors into the eIF4F complex. It has also been shown that oncogenes such as MYC can upregulate the transcription of ribosomal proteins, including the initiation factor protein components of eIF4F, enhancing translational output in the tumor cell. ,
A number of studies, both in animal models and in humans, provide increasing evidence that tumor cells can use the tightly controlled mechanisms regulating mRNA translation as a protective measure to force immune suppression and enable tumor progression. Another example of tumor cells controlling translation is the use of the integrated stress response, which, when activated appears to promote the priority selection of mRNAs with upstream open reading frames (uORFs) in their 5′ UTRs for translation. , It is also important to note that evidence suggests that alterations in the control of translation of mRNAs can occur in cells of the immune system as well as in the dividing tumor cells. Altered translation control occurring in cells of the immune system and in cells of the TME, for example, could cause degradation of neoantigenic peptides in dendritic cells, reduce CTL activity in CD8 + T cells, and, by phosphorylation of eIF4F factors in neutrophils, promote the trafficking of neutrophils to the TME, facilitating tumor metastasis. , ,
The development of actual tumor cell tolerance
From past and current studies using both animal models of cancer and the application of whole genome sequencing (WGC) and RNAseq technologies to dissect the intricacies of the human immune response to cancer, CD8 + T cells have evolved as central to a successful and sustained antitumor response. The development of the T cells critical to this response requires the generation of the large diversity of T-cell receptors (TCRs) required to generate the antigen recognition repertoire through extensive random recombination events. TCRs that could be harmful to the host by recognition of self-peptides under normal conditions enter into a process of self-tolerance to maintain the health of the individual. This, as we know, is a process dependent on thymic development. As a result, there are a number of mechanisms that serve to regulate and limit T-cell activation in the periphery of the immune system to maintain health and nonrecognition of self-antigens. In cancer, much like the immune response to inflammation and infectious disease, the blocking of self-reacting peripheral T cells serves to restrict activation of TCRs and to downregulate T-cell effector functions including clonal expansion and tissue infiltration.
Evidence has shown that the progressing tumor itself can acquire a variety of additional ways to enhance its ability to evade immune control and to even exclude CD8 + T cells from invading the tumor and the TME. Studies suggests that even outside the confines of the tumor and its TME, as, for example, in the tumor’s draining lymph nodes of the peripheral immune system, tumor cells are being protected from immune destruction, a form of tumor cell tolerance. This peripheral immune protection or tumor cell tolerance has been observed when antibody therapies directed against CTLA-4, PD-1, and PD-L1 have shown increased CTL tumor cell destruction, effectively breaking this tolerance in a percentage of patients, resulting in a significant period of therapeutic benefit.
In another example of what might be considered tumor cell tolerance, immunologists have for years marveled at the relationship of the developing fetus to the mother’s immune system and frequently noted similarities between the immune tolerance of the fetus during pregnancy and the ability of the tumor to develop tolerance to the anticancer immune attack in a patient with cancer. It has been noted by cancer biologists that during pregnancy a number of immune-active placental proteins can be identified including the numerous isoforms of pregnancy-specific β-1-glycoproteins (PSGs). , PSGs are produced by the placenta’s syncytiotrophoblast cells and consist of a gene family (PGS-1 to PGS-9 plus PGS-11) located on chromosomes 19.1–19.3. More than 30 PSG isoforms have been identified and PSG proteins are readily measured in the maternal peripheral blood, especially during the third trimester.
In a recent very interesting and innovative study published in 2021, investigators set out to determine whether cancers took advantage of similar preprogrammed immunosuppressive mechanisms that provided a similar form of tolerance for cancer. To ask this question, they determined the level of PSG gene family member expression cancers using The Cancer Genome Atlas (TCGA) of over 10,000 tumors and consisting of 33 cancer types. In normal tissues the expression of PGS genes is rare. The authors found that PGS genes were in fact significantly upregulated in the majority of the tumors they studied; for example, kidney chromophobe carcinoma, 85%; thyroid carcinoma, 74%; adrenocortical carcinoma, 73%; head and neck squamous cell carcinoma, 68%; and cholangiocarcinoma, 67%. They found elevated PGS mRNA expression in at least 10% of all tumors and that PSG-9 was the most commonly upregulated of the PSG gene family in ∼25% of tumors. Perhaps most important, they found that patients with breast cancer, lung mesothelioma, and ovarian tumors identified with higher PGS expression levels had a significantly worse disease outcome. These results highlight the possibility that PGS proteins may play an added important role in generating tumor tolerance. As such, they may prove to be interesting targets to block.
Tumor cell–derived immune suppression
Tumors have been shown to express a variety of immune suppressive membrane-bound determinants and soluble factors released into the TME that can suppress antitumor immunity. Tumor membrane–bound vesicles contain molecules such as FasL, which is recognized by Fas receptors on CD8 + CTL, suppressing CTL function and stimulating the expression and secretion of a number of inhibitory inflammatory cytokines such as interleukin (IL)-10, IL-35, and transforming growth factor beta (TGF-β). This dynamic between antitumor immunity and protumor progression takes place to a large extent within the cellular microenvironment surrounding the tumor and in association with tumor metastases—the collective microenvironment (TME). The tumor and surrounding tissues that comprise the TME include cancer cells, cancer stem cells, cancer-associated fibroblasts, antigen-activated T cells, NK cells, B cells, dendritic cells, endothelial cells, and bone marrow–derived proinflammatory myeloid cells that further differentiate into macrophages, neutrophils, and monocytes. Evidence has been accumulating to place the inflammatory TME in the key position of being an important determinant, if not the determinant, of the effectiveness of both standard anticancer therapies and the newer immunotherapies. For example, conventional therapies (chemotherapy, radiotherapy, and surgery) targeted directly at tumor cell destruction may enhance dendritic cell maturation, antigen presentation, and CD8 + T-cell recruitment, enhancing the antitumor adaptive immune response. In contrast, genomic variants occurring within the cancer cells as well as those variants induced by the cancer in noncancer cells of the TME can cause recruitment and activation of inflammatory cells in the TME, resulting in immunosuppression and tumor progression.
Characteristic of carcinogenesis, the immune system and the chronic inflammation associated with the developing tumor cogenerate a conflicted environment of antitumor suppression and protumor progression. The interaction of these forces is edited in multiple ways during the progression of the disease by the introduction of externally derived cell destructive therapies (chemotherapy, radiation therapy, and surgery); what is even more confounding to this environment of combined pro and con activities is the use of immunotherapies. From the efforts to introduce immune therapeutics into the mainstream of cancer treatment, it is more apparent than ever that there is much work to do to understand how the immune system and the inflammatory response can be effectively joined to treat cancer.
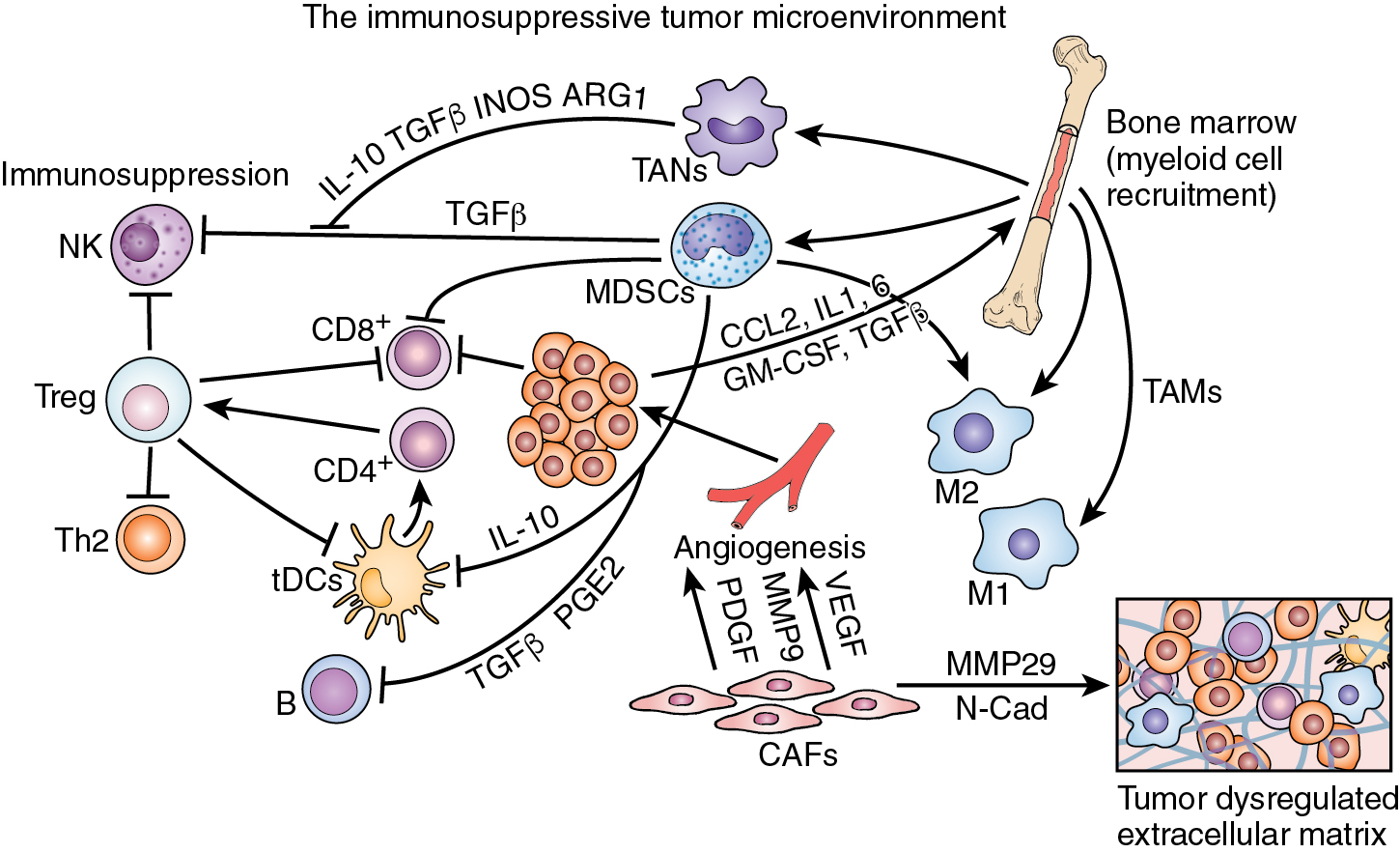
The tumor microenvironment, inflammation, and immune escape
Though much of the discussion in this chapter has centered on the tumor cell itself and its direct interaction with the cells of the immune system, it is critical in any review of the ways in which tumors avoid host immune destruction to recognize that the growing epithelial tumor exists within a desmoplastic complex stroma, the tumor microenvironment (TME). This multicellular supporting environment is a network of stromal cells including cancer-associated fibroblasts (CAFs), vascular endothelial cells, pericytes, various cells of the immune system, and a variety of bone marrow–derived inflammatory cells including mesenchymal cells. As the tumor progresses, it is readily apparent that the TME becomes increasingly supportive of the tumor and increasingly suppressive of antitumor immunity. The cellular heterogeneity of the TME is well recognized, but even more important is the dynamic nature of the environment reflected in the shifting nature of its different cellular composition. This creates changes in cell-to-cell interactions and differential transcriptomics regulating the secretion of specific cytokines and chemokines. This qualitative and quantitative difference in cytokines and chemokines determines differences in inflammatory cell recruitment to the TME. In addition to recruitment of immune cells such as immunosuppressive Tregs, other tumor-promoting cells such as macrophages, monocytes, neutrophils, or innate lymphoid cells are attracted to the TME and to infiltrate the tumor itself. Expansion of specific cell populations within the TME may occur by clonal expansion or by recruitment from the bone marrow ( Fig. 8.4 ).