The Genetic Basis of Antibody Structure
Introduction
In previous chapters we described the enormous diversity of the immune response, focusing on the diversity of antibodies: immunoglobulin (Ig) molecules that are the secreted forms of the antigen-specific receptors found on individual B lymphocytes. In later chapters we will also describe the similar scale of the diversity of antigen-specific T-cell receptors (TCR). Estimates of the number of B and T cells with different antigenic specificities that can be generated in a single individual are very high, in the range of 1011; in other words, every person has the ability to generate 1011 or more different Ig or TCR molecules. However, from the sequencing of the human genome (inherited DNA) that has been conducted over the past few years, we now know that every person has only between 20,000 and 25,000 genes—stretches of DNA that code for a polypeptide (or RNA); the genomes of other mammalian species contain similar numbers of genes.
How do so few genes produce so many different antigen receptor molecules? Studies over the past 30 years have established that there are only a few hundred Ig and TCR genes, but they use a unique rearrangement or recombination strategy to produce millions or more possible protein sequences. In this way, genes, or in effect small gene segments, are combined to generate a single Ig or TCR molecule. A key feature of this rearrangement or recombination mechanism is that Ig and TCR genes change their positions along the DNA sequence, that is, move, during the development of B and T cells, respectively. This was first shown for Ig genes by Susumu Tonegawa, who was awarded the Nobel Prize in 1987 for this work.
In this chapter we discuss how Ig genes are arranged in the genome and the key features of Ig synthesis, from the gene to the protein. We will also discuss the other mechanisms that contribute to the diversity of antigen-specific receptors, and how B cells generate the huge number of antigen-specific receptors that deal with the vast array of pathogens and harmless antigens that confront every individual.
A Brief Review of Nonimmunoglobulin Gene Structure and Gene Expression
Before discussing how an Ig molecule is synthesized, we thought it helpful to review how a typical nonimmunoglobulin gene is organized and how a protein is synthesized from this gene. In Figure 7.1 and below we focus on a gene that codes for a prototypical protein expressed at the cell surface.
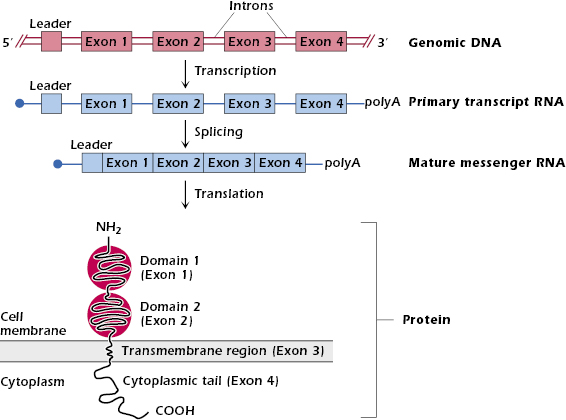
The gene has the characteristic structure shown in the top line of Figure 7.1: exons—sequences of base pairs that are later transcribed into mature messenger RNA (mRNA)—are separated from each other by introns—sequences of base pairs that are removed before a mature mRNA is produced. Typical of genes coding for proteins expressed at the cell surface (or those destined to be secreted or move into intracellular organelles), this gene also has a leader sequence (leader exon) at the 5′ end. This codes for a short sequence of about 10 mainly hydrophobic amino acids—the signal peptide—at the amino (N) terminus of the protein. Not shown in Figure 7.1 are regulatory sequences in front of (that is, 5′) the leader sequence that control gene transcription. These sequences of nucleotides bind proteins called transcription factors that initiate or modulate transcription. Transcription of the gene starts after RNA polymerase has bound to the transcription factor bound to the DNA.
When the gene shown in Figure 7.1 is transcribed into RNA, the entire stretch of DNA (exons plus introns) is transcribed into a primary RNA transcript. A “cap” (a single methylguanosine at the 5′ end) and “tail” (of about 250 adenines at the 3′ end) are added to the ends of the RNA transcript to protect it from degradation. Enzymes then modify the primary RNA transcript by splicing out the introns and bringing together all the exons. This yields a mature mRNA, which leaves the nucleus and is then translated into protein on ribosomes.
When the mRNA for a cell-membrane-associated protein is translated on ribosomes, the signal peptide directs the synthesis of the polypeptide chain to the endoplasmic reticulum. The nascent polypeptide chain is fed from the ribosomes into the interior of endoplasmic reticulum, where the signal peptide is cleaved off. The newly synthesized protein moves from the endoplasmic reticulum into the Golgi apparatus and then to the cell membrane.
Notice in Figure 7.1 the general feature that each exon codes for a discrete region or domain of an individual protein. In the example shown, the four exons code for the two extracellular domains, a transmembrane region, and the cytoplasmic tail.
The surface molecule depicted in Figure 7.1 has some similarities to the structure of a membrane Ig molecule expressed at the surface of a B cell—shown for example in Figure 8.2—especially the extracellular N-terminal domains and transmembrane region. However, membrane Ig also differs in important ways from the structure of the depicted molecule. First, Ig is a four-chain glycoprotein. To make a complete Ig molecule, the newly synthesized individual heavy (H) and light (L) chains must be assembled and glycosylated inside the cell before the four-chain molecule reaches the cell surface. Second, each Ig chain has a very short cytoplasmic tail.
Other molecules involved in the immune response and expressed at the cell surface have different configurations; for example, the carboxy-terminal region can be extracellular and the N-terminus intracellular. Molecules such as CD81, expressed on B cells (see Chapter 8), loop multiple times through the membrane. Still others, such as leukocyte function-associated antigen 1 (LFA-1; CD58) and decay-accelerating factor (DAF; CD55), are completely extracellular; they are linked to the cell surface via a covalent bond to an oligosaccharide, which in turn is bound to a phospholipid (phosphatidylinositol) in the membrane. Thus these molecules are referred to as glycosylphosphatidylinositol (GPI)-linked membrane molecules. (The functions of LFA-1 and DAF are discussed in Chapters 11 and 14, respectively.)
Gene expression. Every diploid cell in the human body contains the same genes as every other cell. The only exceptions are lymphocytes, which, as we discuss shortly, differ from other cells and each other in the actual content of genes coding for their antigen-specific receptor.
Cells within an individual differ from one another because they transcribe and translate different genes. We say that these cells express different patterns of genes. The expression of a specific pattern of genes determines the cell’s function. For example, every cell contains an insulin gene, but only pancreatic β cells express that gene, enabling them to make insulin. Similarly, all cells contain Ig genes; however, only B lymphocytes (and their differentiated form, plasma cells) express Ig genes and therefore synthesize Ig molecules. Like all other cells except B cells, T cells contain Ig genes but do not express them.
The control of gene expression exists at multiple levels, but many factors can either increase (“upregulate”) or decrease (“downregulate”) expression of a particular gene. Controls of gene expression include the activity of transcription factors, the rate of transcription, and the half-life of mRNA. Understanding the mechanisms that regulate gene expression in different cell types is an area of intense research interest. Genes occupy only a very small part (1–2%) of the total genome, and until recently, the rest of the DNA was not considered to have any function. Recent studies, however, have identified multiple gene switches throughout the genome, indicating that several sites in the genome outside of genes play a key role in the control of gene expression.
Genetic Events in Synthesis of Ig Chains
The organization of genes coding for Ig H and L chains shows some similarities to, and some important differences from, the organization of genes for the prototypical cell surface membrane protein we described in the previous section. In addition, as we mentioned earlier in the chapter, genes coding for Ig (as well as genes coding for the TCR) have the unique property of moving—rearranging—during different stages of development of the cell. These properties are described in the sections that follow.
Organization and Rearrangement of Light-Chain Genes
General Features. We describe first the organization and rearrangement of Ig L-chain genes—κ and λ—because these are somewhat simpler than the organization and rearrangement of H-chain genes. We saw in Chapter 4 that each κ and λ L-chain polypeptide consists of two major domains, a variable region (VL)—the approximately 108-residue amino-terminal portion of the L chain—and a constant region (CL).
In the next sections we describe the following:
- Two different gene segments—variable (V) and joining (J) segments—code for the variable region of the L chain.
- A different gene, the C gene, codes for the constant region of the L chain.
- DNA coding for the L chain is cut and V and J gene segments are joined together in the genome by a complex of enzymes called V(D)J recombinase. The joined VJ gene segments, together with the C gene, then synthesize a complete L chain.
The recombinase is called “V(D)J” because, as we describe later in the chapter, H chains use D gene segments as well as V and J segments, and the same recombinase is involved in cutting and joining H chain and L chain DNA. Thus, the mechanism of cutting and joining DNA coding for Ig genes is known as V(D)J recombination.
κ-Chain Synthesis
The top line of Figure 7.2 shows the arrangement of the set of genes coding for human κ chains—referred to as the κ locus (located on chromosome 2)—in the germline, that is, in any cell in the body. There are approximately 40 different Vκ gene segments, arranged linearly and separated from each other by introns, 5 different Jκ segments downstream of the Vκ genes, and a single Cκ gene downstream of the Jκ genes. Each Vκ gene has its own leader sequence, which, for simplicity, has been omitted from the figure. The regions containing Vκ gene segments, Jκ gene segments, and the Cκ gene are separated from each other by long stretches of DNA.
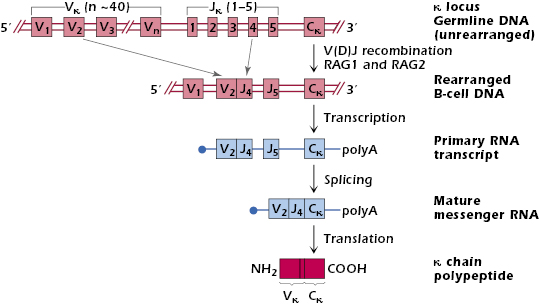
Each Vκ gene segment has the ability to code for the N-terminal 95 amino acids of a κ variable region. Each of the smaller Jκ gene segments can code for the remaining 13 amino acid residues (96–108) of the κ variable region.
Figure 7.2 shows how a κ chain is synthesized by V(D)J recombination. Early in its development in the bone marrow (in the bursa in avian species), each B cell selects one of the Vκ genes from its DNA and physically joins it to one of the Jκ segments; in the B cell shown in the figure, V2 rearranges to J4. The mechanism of selection of V and J genes is unknown, but it is probably random. The stage in B cell development at which this occurs is explained in Chapter 8.
The rearrangement of Vκ to Jκ—and indeed the rearrangement of all Ig and TCR genes—is mediated by the V(D)J recombinase enzyme complex, which cuts and joins different gene segments in DNA. Some of the enzymes of the recombinase complex are involved in the repair of DNA strands in all cells of the body, but two of its components—recombination-activating (RAG)-1 and RAG-2—are expressed exclusively in developing lymphocytes. RAG-1 and RAG-2 are required for the initial cutting of Ig (and TCR) DNA, and so RAG-1 and RAG-2 are critical for the development of B and T cells. Humans whose RAG-1 or RAG-2 gene product is defective lack mature B and T cells; this is one form of severe combined immunodeficiency (SCID) that is described in more detail in Chapter 18. Mice lacking either RAG-1 or RAG-2 genes (“RAG knockout”) are also deficient in both B and T cells.
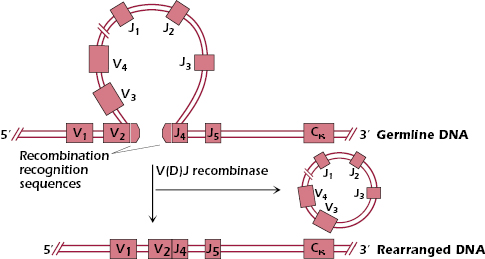
Figure 7.3 shows in more detail how the V(D)J recombinase-mediated cutting and joining of the κ locus DNA occurs. The V(D)J recombinase recognizes recombination recognition sequences that are located at the ends of the V and J gene segments. These recombination recognition sequences are conserved among all V, J (and D) gene segments used by Ig and TCR genes. The recombinase joins together the V2 and J4 gene segments, and the intervening DNA containing V3, V4 etc., is looped, cut out, and ultimately degraded. Note that the rearranged DNA in this early B cell still contains the gene segments V1 and J5 that were not affected by the recombination process.
After the DNA of a cell in the B-cell lineage has been rearranged (V2 to J4), a primary RNA transcript is made, which still contains introns (Figure 7.2). The primary transcript is then spliced to give a mature mRNA in which the Vκ, Jκ, and Cκ exons are brought together and the introns and additional J exons are removed. In the rough endoplasmic reticulum, this mRNA is translated into a complete κ polypeptide chain. The κ chain then moves into the lumen of the endoplasmic reticulum where the signal peptide (encoded by the leader sequence) is cleaved off, and the κ chain can now associate with a newly synthesized H chain to form an Ig molecule.
We explain in more detail below that every B cell has two sets of chromosomes—one set from each parent—and hence two κ loci. The rearrangement mechanism we have described occurs in a sequence. The cell first tries rearrangement at one of the two κ loci. If this results in a functional Ig L chain (a “productive” rearrangement), rearrangement does not occur at either the other κ locus or the two λ loci; they remain in germline configuration. If the result of the rearrangement at the first κ locus is nonproductive, however, rearrangement at the other κ locus, and then the λ loci is tried sequentially.
λ-Chain Synthesis
The λ locus is found on chromosome 22 in the human, that is, on a chromosome distinct from the κ and the H-chain loci. Rearrangement of the λ locus occurs when rearrangement at both κ loci has been unsuccessful (nonproductive). The mechanism of λ locus rearrangement is similar to what we just described for κ chains. The synthesis of λ chains uses V and J gene segments and rearrangement of DNA mediated by V(D)J recombinase: one of about 30 Vλ gene segments is joined to one of the 4 Jλ gene segments. Each Vλ gene segment codes for the N-terminal region of a λ variable region, and each Jλ segment codes for the remaining 13 amino acids of the λ variable region. The organization of the λ locus is slightly different from that of the κ locus, which contains only one Cκ gene: each Jλ is associated with a different Cλ gene. Thus, each λ chain will have one of four possible Cλ regions. Note though that these different Cλ have similar functions, so they should not be considered the equivalent of different H-chain constant (CH) regions, which we described in Chapter 5 as having different effector functions.
Once the Vλ and Jλ segments have rearranged in a particular B cell, the steps in the synthesis of a λ chain polypeptide are very similar to those already described for the synthesis of a κ-chain polypeptide (see Figure 7.2).
Organization and Rearrangement of Heavy-Chain Genes
Figure 7.4 shows the organization of genes coding for immunoglobulin H chains—the H-chain locus. The human H chain locus is found on chromosome 14, distinct from either L-chain locus. The figure illustrates the similarities and differences of this locus with the L-chain loci. The first major difference is that the three different gene segments VH, DH, and JH code for the variable region of the H chain. Thus, in addition to VH and JH segments, genes coding for the variable region of an H chain also use a diversity (D) segment. The D and J segments code for amino acid sequences in the third hypervariable region or complementarity-determining region
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree