Daniel W. Fitzgerald, Timothy R. Sterling, David W. Haas Keywords acid-fast bacilli (AFB); aminoglycoside; bacillus Calmette-Guérin (BCG); bedaquiline; directly observed therapy (DOT); ethambutol; extensively drug-resistant tuberculosis (XDR-TB); fluoroquinolone; isoniazid (INH); multidrug-resistant tuberculosis (MDR-TB); Mycobacterium bovis; N95 mask; pyrazinamide; rifampin (RMP); tuberculosis
Mycobacterium tuberculosis
The term tuberculosis describes a broad range of clinical illnesses caused by Mycobacterium tuberculosis (or, less commonly, Mycobacterium bovis). It is second only to human immunodeficiency virus (HIV) as a cause of death worldwide resulting from a single infectious agent. In 1993 the World Health Organization (WHO) declared tuberculosis a global public health emergency, and tuberculosis continues to be an immense global public health problem. Tuberculosis can affect virtually every organ, most importantly the lungs, and is typically associated with granuloma formation.
History
There is evidence of spinal tuberculosis in Neolithic, pre-Columbian, and early Egyptian remains. However, tuberculosis did not become a major problem until the Industrial Revolution, when crowded living conditions favored its spread. In the 17th and 18th centuries, tuberculosis caused one fourth of all adult deaths in Europe. Before antimicrobial agents became available, the cornerstone of treatment was rest in the open air in specialized sanatoria. Sanatorium regimens probably benefitted some cases diagnosed before cavitation but had little impact on cavitary disease. When it became clear that cavitation was the pivotal event in progressive pulmonary tuberculosis, most special therapies focused on cavity closure.
The modern era of tuberculosis began in 1946 with demonstration of the efficacy of streptomycin (STM). In 1952, the availability of isoniazid (INH) made tuberculosis curable in most patients, and the addition of rifampin (RMP) in 1970 allowed for even more effective combination therapy. With drug coverage, it became possible to successfully resect tuberculous tissue, but with drug treatment, resection was rarely necessary. Bed rest and collapse therapy added nothing to chemotherapy; treated patients rapidly became noninfectious; and specialized sanatoria ultimately disappeared. The duration of chemotherapy progressively decreased from approximately 2 years before the availability of RMP, to 9 months with INH plus RMP, and to 6 months using multidrug therapy including INH, RMP, and pyrazinamide (PZA). With INH it also became practical to treat asymptomatic people believed to harbor tubercle bacilli based on positive tuberculin tests.
In the United States, reported cases of tuberculosis had declined nearly every year since accurate statistics became available. However, in 1985 case rates began increasing, driven largely by HIV infection. Tuberculosis control programs in some large cities were not equipped to manage this emerging problem. The often-interrelated factors of illicit drug use, homelessness, and HIV infection predispose to reactivation of remote tuberculosis, to the acquisition and spread of new disease, and, because of irregular adherence to drug therapy, to the development and spread of drug-resistant strains. Epidemics involving strains that were resistant to at least INH and RMP (i.e., multidrug-resistant [MDR]) emerged in these populations and spread to HIV-negative persons, including health care workers. Many outbreaks were caused by the Beijing strain, with “strain W” dominating in New York City.
Treatment programs failed because of drug resistance, patient nonadherence, and nosocomial transmission of M. tuberculosis. Since 1992, however, tuberculosis incidence rates in the United States have again declined and in 2012 reached the lowest in history.1 This attests to the success that can be achieved with intensified diagnostic, treatment, and prevention efforts, and with control of HIV-induced immunocompromise by antiretroviral therapy.
The global situation has, unfortunately, not been as successful. The HIV pandemic fueled increased tuberculosis case rates in resource-limited countries worldwide, especially in sub-Saharan Africa. Scant resources and fragile infrastructure, together with a high prevalence of HIV infection and acquired immunodeficiency syndrome (AIDS), has driven the global burden of tuberculosis higher. As in the United States, MDR tuberculosis (MDR-TB) emerged and spread. In response, WHO has worked diligently to expand tuberculosis treatment services, including directly observed therapy, short-course (DOTS) programs. In parallel, second-line medications, including fluoroquinolones, were made increasingly available worldwide, with a functional DOTS program being a prerequisite for access to discounted drug pricing.2
With widespread use of second-line agents, selection for M. tuberculosis resistant to both first- and second-line drugs was inevitable. Extensively drug-resistant tuberculosis (XDR-TB), defined as resistance to at least INH, RMP, a fluoroquinolone, and a second-line injectable drug (kanamycin, capreomycin, amikacin) first occurred as early as 2001 in Kwazulu-Natal, South Africa,3,4 and by 2013 it had been reported in at least 84 countries worldwide.5 Although application of established tuberculosis public health principles, supported by ample funding, may ultimately control this dire situation,6 the immensity of this challenge is daunting.
Microbiology
The M. tuberculosis complex comprises at least seven species in the genus Mycobacterium, family Mycobacteriaceae, and order Actinomycetales that are causes of human tuberculosis and zoonotic disease. The M. tuberculosis complex species share 99.9% sequence identity and likely evolved from a single clonal ancestor.7 The species M. tuberculosis sensu stricto causes the vast majority of human tuberculosis worldwide. Mycobacterium africanum causes human tuberculosis in West Africa, where it accounts for up to 50% of cases.8 Mycobacterium canetti is an extremely rare cause of human tuberculosis in the Horn of Eastern Africa. M. bovis causes disease in cattle and spreads to humans through animal contact and consumption of unpasteurized milk. An investigation of six tuberculosis cases in the United Kingdom demonstrates that M. bovis can rarely be transmitted from human to human.9 Mycobacterium caprae, another cattle pathogen, Mycobacterium microti, a pathogen for rodents, and Mycobacterium pinnipedii, a pathogen for seals, have been reported to cause zoonotic tuberculosis in humans.
Advances in genetic analysis, including whole-genome sequencing, have shed new light on the phylogenetics of the M. tuberculosis complex.10 These studies show that M. tuberculosis sensu stricto and M. africanum, the two predominate causes of human disease, can be further divided into six phylogenetic lineages, including Indo-Oceanic, East Asian, East African-Indian, Euro-American, West African I, and West African II. This is a rapidly changing field of study, with multiple nomenclatures in use. For example, the East Asian lineage is commonly called the Beijing strain. There is some evidence that different lineages vary in virulence, transmissibility, or ability to acquire drug resistance, but further research is needed to clarify the clinical importance of such differences.
Humans are the only reservoir for the species M. tuberculosis, although many animals are susceptible to infection.11 Some have postulated that an ancient ancestor of M. tuberculosis infected hominids in East Africa 3 million years ago and has since coevolved with its human host.12 M. tuberculosis is an aerobic, non–spore-forming, nonmotile bacillus with a high cell wall content of high-molecular-weight lipids. Growth is slow, the generation time being 15 to 20 hours, compared with much less than 1 hour for most common bacterial pathogens, and visible growth takes from 3 to 8 weeks on solid media. The organism tends to grow in parallel groups, producing the colony characteristic of serpentine cording. In radical contrast to other bacteria, a very large portion of M. tuberculosis genes encode enzymes involved in lipogenesis and lipolysis.
A wide spectrum of laboratory techniques has been developed to diagnose active tuberculosis. No single test is perfect, and, unfortunately, some diagnostics on which clinicians still rely were developed over 100 years ago. Advantages and limitations of various methods are presented in Table 251-1.
Acid-Fast Staining
The term acid-fast bacilli is practically synonymous with mycobacteria, although Nocardia and some other organisms are variably acid fast. In the Ziehl-Neelsen stain, a fixed smear covered with carbol-fuchsin is heated, rinsed, decolorized with acid-alcohol, and counterstained with methylene blue. The Kinyoun stain is modified to make heating unnecessary. The organisms appear as slightly bent, beaded rods 2 to 4 µm long and 0.2 to 5 µm wide. In sputum they often lie parallel or two organisms adhere at one end to form a V. An estimated 10,000 organisms/mL of sputum are required for smear positivity, and detection of at least 10 organisms on a slide is optimal; a single organism on a slide is highly suggestive. The sensitivity of sputum acid-fast bacillus smear when compared with culture is approximately 60%.13 Sensitivity is significantly lower with noncavitary disease and HIV infection. Sensitivity increases by approximately 10% with the collection of a second sputum sample, and 2% with a third.14 Sputum processing with bleach and concentration before acid-fast staining also increases sensitivity.15 Most laboratories in the United States now use a fluorochrome stain with phenolic auramine or auramine-rhodamine, a slightly modified acid-alcohol decolorization step, and potassium permanganate counterstaining. Because the mycobacteria are easily seen with a 20× or 40× low-magnification objective, fluorescence microscopy requires less technician time and may increase the sensitivity over conventional acid-fast bacillus smears.16 Advances in ultrabright light-emitting diode (LED) microscopes make the technology more robust for use in resource-poor settings.17 In 2011, the WHO recommended that LED microscopy replace conventional fluorescence microscopy and that it be phased in as an alternative for conventional Ziehl-Neelsen microscopy.18
Any biologic fluid or material can be examined directly (e.g., pleural fluid, cerebrospinal fluid [CSF], urine, gastric lavage fluid), although thin fluids are best examined after sedimentation by centrifugation. Positive smears from concentrated gastric aspiration material are usually due to M. tuberculosis and are especially important in young children from whom sputum collection may not be possible.
Culture Methods for M. tuberculosis
Culture is the gold standard for detecting mycobacteria in clinical specimens. Samples of sputum or tissue require initial decontamination to remove fast-growing nonmycobacterial organisms and liquefaction to allow access of decontaminants to nonmycobacterial organisms and media nutrients to surviving mycobacteria. Decontamination-liquefaction is most commonly done using N-acetyl-l-cysteine as a mucolytic in 1% sodium hydroxide solution. Mycobacteria are relatively protected during this procedure by a fatty acid–rich cell wall. However, normally sterile tissues or fluids such as CSF or pleural fluid should not be decontaminated, because some loss of mycobacterial viability does occur. The sample is then neutralized and centrifuged, and the sediment is inoculated onto media.
Three types of media may be used for culture of mycobacteria: solid egg-based (e.g., Lowenstein-Jensen), solid agar-based (e.g., Middlebrook 7H11), and liquid broth (e.g., Middlebrook 7H12). Media are made selective for mycobacteria by adding antibiotics. Nonselective media, on which growth is more rapid, are available. Growth is more rapid in 5% to 10% carbon dioxide. Liquid broth cultures require 1 to 3 weeks of incubation for detection of organisms, as compared with solid media, which require 3 to 8 weeks. However, solid media allow examination of colony morphology, detection of mixed cultures, and quantification of growth. Further, occasional strains of mycobacteria may only grow on solid media. For these reasons, experts suggest using liquid and solid media in conjunction, with inoculation of at least one solid medium culture.19
Commercial automated liquid broth systems greatly facilitate mycobacterial culture. They monitor mycobacterial growth by detection of CO2 production or O2 consumption through radiometric, fluorometric, or colorimetric indicators. The BACTEC mycobacterial growth indicator tube (MGIT) system (Becton Dickinson Microbiology Systems, Sparks, MD), which detects growth in 1 to 3 weeks by a fluorometric method, is widely used.20,21
A noncommercial liquid broth assay has been developed in which mycobacteria are cultured in liquid media on a multi-well plate and then examined microscopically for characteristic serpentine cording. The addition of antimicrobial agents to the media allows drug susceptibility testing to be performed simultaneously. This microscopic-observation drug-susceptibility (MODS) assay yields results in 7 to 10 days with sensitivity and specificity similar to commercially available liquid broth systems.22 This promising, inexpensive, and rapid method for culture and drug susceptibility testing will need to be standardized before it is widely adopted.
Nucleic Acid Amplification
Nucleic acid amplification tests (NAATs) offer another technique for the direct detection of M. tuberculosis in clinical specimens. The GeneXpert MTB/RIF (Cepheid, Sunnyvale, CA) is an automated molecular test for detection of M. tuberculosis with sensitivity and specificity that approaches culture.23 The test is simple to perform and gives results in 100 minutes. It uses real-time polymerase chain reaction (PCR) amplification of an M. tuberculosis gene for detection. Analysis of three sputum samples with GeneXpert MTB/RIF had an overall sensitivity of 98% compared with gold standard culture testing. Sensitivity was 99.8% for smear- and culture-positive cases and 90.2% for smear-negative culture-positive cases.24 This assay simultaneously detects rifampin resistance (see “Drug Susceptibility Testing,” later). The assay can also be used on nonrespiratory samples (e.g., pleural fluid, CSF, urine, fine-needle aspirates) on which it has approximately 70% sensitivity compared with culture. More studies are needed to define its role in the diagnosis in extrapulmonary disease. The assay has been endorsed by the WHO and is rapidly being deployed to countries with high rates of endemic tuberculosis. Studies are underway to gain U.S. Food and Drug Administration (FDA) approval.
At least two older NAATs are commercially available and FDA approved: the Amplified M. tuberculosis Direct Test (Gen-Probe, San Diego CA), which targets ribosomal RNA, and the AMPLICOR M. tuberculosis Test (Roche Diagnostic Systems, Basel, Switzerland), which targets DNA. The sensitivity of these NAATs is intermediate between acid-fast staining and culture. For smear-positive specimens, the sensitivity and specificity exceed 95%. For smear-negative cases, sensitivity has ranged from 40% to 77% and the specificity remains over 95%.25
Nucleic acid amplification complements but does not replace clinical judgment, acid-fast smear, and culture in the diagnosis of tuberculosis.26 In sputum acid-fast smear-positive individuals, positive nucleic acid amplification indicates the presence of M. tuberculosis complex and confirms active tuberculosis. When there is a high clinical index of suspicion for pulmonary tuberculosis but with a negative acid-fast smear, a positive NAAT is highly predictive of tuberculosis and allows early initiation of therapy. NAATs perform less well when the clinical index of suspicion for tuberculosis is low, in which case the frequency of false-positive tests may approach that of true positives.27
Detection of Mycobacterial Antigens in Urine
Detection of M. tuberculosis antigens in urine may have some utility in diagnosing tuberculosis in patients with advanced AIDS, particularly in countries where coinfection is highly prevalent. The most studied target antigen is the cell wall lipopolysaccharide lipoarabinomannan (LAM). Among patients with CD4 T-cell counts less than 50 cells/mm3, sensitivity of urinary LAM testing has ranged from 56% to 85%, exceeding the sensitivity of sputum smear microscopy, and with specificity exceeding 88%.28 The combination of microscopy and urinary LAM testing provides incremental diagnostic sensitivity. Sensitivity for tuberculosis in HIV-uninfected persons is less than 25%. A urine dipstick format could make this assay amenable to point-of-care use.28
Speciation of Mycobacteria
Once mycobacteria have been identified in a clinical specimen, speciation is necessary for clinical diagnosis and epidemiologic investigation. For example, speciation may be important in immunocompromised patients at risk for nontuberculous mycobacterial infection, in localities where M. bovis transmission from animals to humans is possible, or in bladder cancer patients receiving bacillus Calmette-Guérin (BCG) immune stimulatory therapy. Speciation generally involves two steps: first, mycobacteria are identified as members of the M. tuberculosis complex (M. tuberculosis, M. bovis, M. africanum, M. microti, M. canetti, M. caprae, and M. pinnipedii) or as mycobacteria species other than tuberculosis (MOTT). Subsequently, if necessary, mycobacteria can be speciated within the M. tuberculosis complex. NAATs, growth in selective antibiotic media, nucleic acid probes, and high-performance liquid chromatography are all used to place mycobacteria within the M. tuberculosis complex.29 Each has advantages and limitations that are detailed in Table 251-1. Identifying individual species within the M. tuberculosis complex is more challenging. M. tuberculosis grows slowly, lacks pigment, produces niacin, reduces nitrates, has weak catalase activity that is lost by heating to 68° C at pH 7.0, does not demonstrate monodrug resistance to PZA, is resistant to thiophen-2-carboxylic acid hydrazide, and prefers aerophilic conditions. Other members of the complex show different patterns on these tests. PCR-based genomic deletion analysis, which capitalizes on available complete genome sequence for M. tuberculosis, can also identify species within the M. tuberculosis complex.30
Genotyping of M. tuberculosis
Characterizing the particular strain of M. tuberculosis is important for epidemiologic purposes, such as tracing transmission from person to person, distinguishing exogenous reinfection from endogenous reactivation in cases of recurrent tuberculosis, and identifying laboratory cross-contamination of cultures. The Centers for Disease Control and Prevention (CDC) offers free strain typing through the National Tuberculosis Genotyping Service based on three standardized typing methods: restriction fragment length polymorphism (RFLP) analysis, spacer oligonucleotide typing (spoligotyping), and mycobacterial interspersed repetitive unit (MIRU) analysis.31 In RFLP analysis, DNA cleavage fragments generated using pvuII are separated by electrophoresis and visualized using a probe to a repetitive DNA sequence, insertion sequence (IS) 6110. Because numerous copies of IS6110 are present at variable chromosomal locations in most M. tuberculosis isolates, identical RFLP patterns represent the same strain. Spoligotyping detects variability in the direct repeat (DR) region in M. tuberculosis genome. There are direct repeats of a conserved 36-bp sequence, separated by multiple spacer sequences. Different M. tuberculosis strains are distinguished by the presence or absence of 43 unique spacers. Each possible combination of spacers is designated by a unique numerical code that identifies a unique spoligotype strain. In MIRU analysis, 12 different DNA sequences, each of which can be tandemly repeated in the M. tuberculosis genome, are analyzed. The number of tandem repeats for each of the 12 sequences (or loci) is determined by PCR to create a 12-number code. Each code corresponds to a unique MIRU strain.
Drug Susceptibility Testing
Testing of M. tuberculosis isolates for drug susceptibility is important to guide therapy. In the United States, the agar proportion method is most commonly used. The absolute concentration method and resistance ratio method are used less commonly. The agar proportion method compares growth of appropriately diluted inocula on drug-containing media to growth on drug-free media and is reported as proportion resistant. For most drugs, resistance is significant when growth on drug-containing media exceeds 1% of control; 6% to 10% resistance or more indicates that the drug will add nothing to multidrug therapy. Liquid broth systems, such as BACTEC, can also be used and provide results in 5 to 14 days, but they do not give the proportion of resistant organisms.32,33
As noted earlier, the MODS method shows promise as an inexpensive and rapid method for culture and drug susceptibility testing.22
Molecular testing to detect chromosomal mutations associated with mycobacterial drug resistance is an exciting development.24,34–37 Most useful are tests for RMP resistance, which predicts poor treatment outcomes and is a surrogate marker for MDR-TB. Assays detect mutations in the 81-base-pair rpoB gene, which encodes the β-subunit of RNA polymerase and correlates with greater than 96% of RMP resistance. Resistance to INH is more complex and is encoded by multiple genes, including the catalase peroxidase gene katG, the inhA gene involved in fatty acid biosynthesis, the ahpC gene, the oxyR gene, and the kasA gene.38 Mutations associated with resistance to PZA, ethambutol (EMB), STM, and fluoroquinolones have also been identified.
The GeneXpert MTB/RIF real-time assay described earlier correctly identifies 98% of cases that are rifampin resistant by culture techniques and 98% of rifampin-sensitive cases. The drug resistance assay is performed on sputum simultaneously with detection of organisms, and results are available within 2 hours. Two line probe assays are also commercially available: the INNO-LiPA Rif. TB kit (Innogenetics, Zwijndrecht, Belgium), which detects resistance to RMP on culture isolates, and the Genotype MTBDRplus assay (Hain Lifescience, Nehren, Germany), which detects resistance to INH and RMP on culture isolates and sputum samples.39,40 The WHO has recommended widespread use of these molecular assays.41
Epidemiology
General Considerations
M. tuberculosis infects one third of the world’s population and causes 8.7 million new cases of tuberculosis and approximately 1.4 million deaths each year.5 Tuberculosis is second only to HIV as a cause of death worldwide resulting from a single infectious agent.42 Immunocompromise due to HIV infection is a risk factor for tuberculosis, and 0.4 million tuberculosis deaths are in HIV-infected individuals. Drug-resistant tuberculosis is emerging globally, with approximately 630,000 million prevalent MDR-TB cases in 2011. Two factors essential for the rapid spread of M. tuberculosis are crowded living conditions and a population with little native resistance. In the 19th century, tuberculosis caused more than one fourth of all adult deaths in Europe, eliminating those with the least native resistance. A downward trend had been established before the turn of the 20th century. Epidemiologists once believed the disease would eventually disappear based on the assumptions that 1 in 20 infections result in active cavitary disease of the lung (i.e., become contagious). Thus, each cavitary case would have to infect 20 persons to maintain case rates.43 In Holland in the early 1900s, one infectious case produced only 13 new infections.43 The annual decrement in mortality and morbidity from tuberculosis was approximately 5% in developed countries due to progressively higher natural residual resistance in those who survived infection and to living conditions less conducive to airborne spread. This rate of decline approximately doubled after chemotherapy became widespread.
In the 1980s and 1990s, the high incidence of tuberculosis in Africa, Asia, Eastern Europe, and South America, the HIV co-epidemic, and burgeoning MDR-TB demonstrated that predictions of the disappearance of tuberculosis were premature and the WHO declared tuberculosis a global public health emergency. Since then, tremendous gains have been made in reducing the number of new tuberculosis cases, now in decline for the past several years.
Recent Morbidity and Mortality Trends
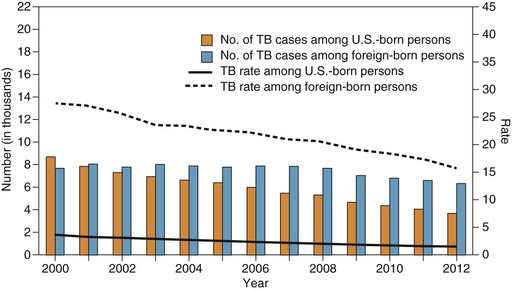
In the United States, the incidence of tuberculosis has decreased each year for 20 consecutive years, from 10.5 reported cases per 100,000 population in 1992 to 3.2 cases per 100,000 population in 2012, the lowest in recorded history.1 In the United States the tuberculosis rate in foreign-born persons is 11 times higher than in U.S.-born persons, and foreign-born persons now account for almost two thirds of reported cases in the United States (Fig. 251-1). This reflects increased immigration from high-prevalence countries, especially Mexico, the Philippines, Vietnam, India, and China, from which immigrants account for more than half of the foreign-born tuberculosis cases.44 Strain genotyping suggests that tuberculosis among foreign-born persons is from reactivation of latent infection acquired before arrival in the United States. Further, the risk declines with duration of residency, also suggesting that most infections were acquired before immigration.45
Tuberculosis has also become concentrated in certain ethnic/racial minorities and medically underserved populations, often occurring in contact-based microepidemics. In 2012, rates among blacks and Hispanics were 6 to 7 times higher than in non-Hispanic whites; the rate in Asians was 25 times higher than in whites. A number of outbreaks have affected the urban poor, alcoholics, injection drug users, the homeless, migrant farm workers, and prison inmates.
The age distribution of tuberculosis reflects the degree of ongoing transmission in a given population. Disease in the elderly is generally due to reactivation of infection acquired in the remote past, whereas tuberculosis in young children indicates ongoing active transmission in the community. In this regard, 75% of childhood cases in the United States had tuberculosis exposure through foreign-born parents or prior residence outside the United States.46
Tuberculosis in the United States is most frequent in geographic regions and demographic groups where AIDS is prevalent, notably in urban blacks and Hispanics between 25 and 45 years of age.47 Persons with active tuberculosis are more frequently HIV positive than is the general population. Approximately 8% of all tuberculosis cases in the United States are in HIV-infected persons.1
Despite a predominantly urban epidemiology, large tuberculosis outbreaks have also affected small communities.48,49 One well-characterized outbreak began in 1988 in a coastal Maine village where tuberculosis had not been reported in the previous 3 years.49 A shipyard worker with cavitary tuberculosis was the source of 21 subsequent active cases and 697 new tuberculous infections. In retrospect, the source case had repeatedly sought medical attention for cough, sore throat, and hoarseness during 8 months before tuberculosis was diagnosed and treated. This report highlights the need for vigilance among all segments of the population, not just those known to have high tuberculosis case rates.
The prevalence of positive TSTs in U.S. Navy recruits offers insights into trends of latent tuberculosis infection over time and risk factors for infection. From 1958 through 1969, more than 1 million U.S. Navy recruits received TSTs and 5.2% were positive. In the 1980s and 1990s, the rate of tuberculin reactivity dropped to approximately 1.5% of new Navy recruits. The prevalence was greater in blacks (5%), Hispanics (5%), and Asian/Pacific Islanders (26%) than in whites (0.8%). Tuberculin positivity was more than 10-fold more prevalent among foreign-born recruits.50–52 In a survey of noninstitutionalized civilians in the United States, the prevalence of tuberculin reactivity among persons from 25 to 74 years of age decreased from 14.4% in 1972 to 5.6% in 2000.53
Immigrants for the most part retain the tuberculin positivity and tuberculosis rates of their country of origin (Table 251-2).54,55 Other groups, such as injection drug users, patients with end-stage renal disease or diabetes, health care workers in endemic countries, residents of institutions for the homeless, and, to a lesser degree, nursing home residents, demonstrate morbidity rates greatly in excess of the general population.56–58,59–64
TABLE 251-2
Reported Tuberculosis Case Rates in Immigrants According to Country of Birth: Stratified by Time since Entry into the United States
| COUNTRY | CRUDE RATE (PER 100,000 PERSON-YEARS)* | |
| U.S. Entry ≤2 Years | U.S. Entry >2 Years | |
| Somalia | 889 | 179 |
| Ethiopia and Eritrea | 562 | 82 |
| Vietnam | 319 | 47 |
| Cambodia | 307 | 65 |
| Philippines | 283 | 38 |
| Ecuador | 194 | 31 |
| Haiti | 189 | 40 |
| Honduras | 177 | 28 |
| Peru | 159 | 32 |
| Guatemala | 111 | 21 |
| India | 106 | 33 |
| China | 74 | 26 |
| El Salvador | 73 | 11 |
| Mexico | 52 | 14 |
| Korea | 40 | 19 |
| All foreign-born | 75 | 16 |
* Data provided for the 15 most commonly reported countries of birth.
Modified from Cain KP, Benoit SR, Winston CA, et al. Tuberculosis among foreign born persons in the United States. JAMA. 2008;300;405-412.
On a global scale, tuberculosis has a devastating impact in developing nations, with 10 countries accounting for nearly 70% of all prevalent cases (Table 251-3).5 In 1993, the WHO declared tuberculosis a global public health emergency and intensified major initiatives to address the problem. An important aspect of this strategy is supervised treatment, which may include DOTS.65 Over the past few years, the WHO reports decreasing incidence rates of tuberculosis in all six regions of the world, including Southeast Asia, the Western Pacific, and Africa. In China, access to DOT was rapidly expanded from 31% of tuberculosis cases in 2001 to 80% in 2005.66 Similar progress has also been made in India, the country with the greatest number of tuberculosis cases.67,68
TABLE 251-3
Estimated Burden of Tuberculosis Cases among Countries That Account for 70% of Cases Worldwide, 2011
| COUNTRY | CASES | RATE PER 100,000 POPULATION |
| India | 3,100,000 | 249 |
| China | 1,400,000 | 104 |
| Indonesia | 680,000 | 281 |
| Pakistan | 620,000 | 350 |
| Bangladesh | 620,000 | 411 |
| Philippines | 460,000 | 484 |
| South Africa | 390,000 | 768 |
| Democratic Republic of Congo | 350,000 | 512 |
| Vietnam | 290,000 | 323 |
| Nigeria | 280,000 | 171 |
| Myanmar | 240,000 | 506 |
| Ethiopia | 200,000 | 237 |
| Russian Federation | 180,000 | 124 |
Modified from World Health Organization. Global Tuberculosis Report 2012. Available at http://www.who.int/tb/publications/global_report/gtbr12_main.pdf.
Over 30 million persons worldwide are now living with HIV infection/AIDS.69 The potential for continued interaction between AIDS and tuberculosis is therefore immense. In some developing countries where most persons harbor tubercle bacilli before adulthood, the prevalence of HIV infection becomes the only determinant of coinfection. The situation is worst in sub-Saharan Africa, where the incidence of tuberculosis has risen in parallel with the incidence of HIV infection. Between 1990 and 2005, the incidence of tuberculosis more than doubled from 149 to 343 per 100,000 population. In 2011, 80% of the world’s HIV-positive tuberculosis cases were in Africa. It is estimated that 30% to 40% of all AIDS deaths in Africa are from tuberculosis. The scale-up of antiretroviral therapy in Africa and the integration of HIV infection and tuberculosis programs offers promise to decrease the impact of these co-epidemics.
Drug-Resistant Tuberculosis
Drug-resistant tuberculosis poses an immense challenge for tuberculosis control. Resistance to antituberculous agents can be either primary, that is, present before initiating therapy and due to transmission of a drug-resistant M. tuberculosis strain, or secondary, indicating emergence of resistance after having received antituberculosis therapy. Risk factors for infection with drug-resistant tuberculosis are listed in Table 251-4. Strains resistant to at least INH and RMP are defined as MDR. Strains resistant to at least INH, RMP, a fluoroquinolone, and an aminoglycoside are defined as XDR.70
TABLE 251-4
Epidemiologic Circumstances in Which an Exposed Person Is at Increased Risk for Infection with Drug-Resistant Mycobacterium tuberculosis*