Fig. 7.1
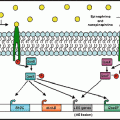
Nutritional Immunity: a competition between mammalian iron sequestration and bacterial iron acquisition that influences bacterial growth proficiency in the mammalian host. Transferrin and other host iron binding proteins sequester iron, thereby preventing growth of microorganisms at systemic locations. Members of the Enterobacteriaceae family including Salmonella enterica produce enterochelin, a siderophore with a high affinity for iron. The bioavailability of enterochelin for bacterial iron acquisition is reduced by the mammalian siderocalin as well as the affinity of enterochelin for lipid membranes. Salmonella glucosylation of enterochelin to salmochelin by IroB reduces both membrane affinity and siderocalin binding. The increased bioavailability of salmochelin compared to enterochelin facilitates iron acquisition from transferrin to support pathogen growth at systemic sites. Epinephrine/norepinephrine are not required for bacterial iron acquisition in vivo. However, due to their siderophore-like properties, Epi/NE accelerate bacterial iron acquisition and therefore when present can enhance pathogen growth in iron-limited environments.(Illustration by Michael Marti)
The ability of NE to enhance the growth of S. Typhimurium in serum-SAPI minimal medium (Freestone et al. 1999; Williams et al. 2006; Bearson et al. 2008) suggests that additional biosynthetic pathways would be modulated besides the iron utilization and transport genes. Transcriptional analysis using microarrays on S. Typhimurium grown in serum-SAPI medium containing 2 mM NE revealed that NE exposure increases transcription of genes involved in amino acid biosynthesis, cofactor biosynthesis, central intermediary metabolism, energy metabolism, and synthesis of transport and binding proteins (Bearson et al. 2008). Thus, to take advantage of the increased availability of iron provided by NE in serum-SAPI minimal medium, S. Typhimurium modulates the biosynthesis of multiple cellular pathways to increase growth rate.
7.2.5 SPI-1 and Invasion
Genes encoded in Salmonella pathogenicity island 1 (SPI-1) are important for invasion of eukaryotic cells. Moreira and Sperandio demonstrated a 1.5-fold increase in the invasion of HeLa cells by wild-type S. Typhimurium in the presence of 50 μM Epi compared to the absence of the catecholamine (Moreira and Sperandio 2012). For quantitative real-time RT-PCR, cultures were grown aerobically to late log phase (O.D.600 = 1.0) in LB medium with or without 50 μM NE or Epi (Moreira et al. 2010; Moreira and Sperandio 2012). Transcription of invF and sopB encoded in SPI-1 were increased >15-fold in LB medium in the presence of 50 μM NE compared to the absence of NE, whereas 50 μM Epi increased sopB and sipA transcription ~twofold in LB compared to the absence of Epi. In contrast to Moreira and Sperandio, the microarray analysis by Karavolos et al. following 30 min of exposure to 50 μM Epi (adrenaline) in LB medium during late log phase (O.D.600 ~ 1.0) indicated that invF (encoding an invasion protein) was down-regulated (Karavolos et al. 2008). Pullinger et al. using a prgH–gfp reporter strain in serum-SAPI medium with and without 5 mM NE indicated that the transcription of prgH was low and not differentially expressed (Pullinger et al. 2010a).
7.2.6 Salmonella Pathogenicity Island 2 (SPI-2) and SPI-2 Effectors
Intracellular survival of Salmonella in epithelial cells and macrophages requires genes encoded within Salmonella pathogenicity island 2 (SPI-2) and additional SPI-2 effectors located outside of SPI-2 (Figueira and Holden 2012). Transcription of ssaG from a gfp transcriptional fusion was significantly reduced in S. Typhimurium cultures grown 16 h in serum-SAPI medium containing 5 mM NE compared to medium without NE (Pullinger et al. 2010a). The authors also demonstrated a reduction in ssaG expression in the presence of 25 μM Fe (III) and noted that this reduction was similar to the effect of NE. Growth of wild-type S. Typhimurium in LB medium to late log in the presence of 50 μM NE enhanced transcription of sifA greater than fivefold compared to LB medium in the absence of NE (Moreira et al. 2010; Rasko et al. 2008). A similar threefold induction of transcription was seen for cultures grown to late log in N-minimal medium pH 4.5 in response to 50 μM Epi exposure (Moreira and Sperandio 2012).
7.2.7 PmrAB Regulon
Members of the PmrAB regulon (pmrFGHIJ) were down-regulated in the presence of Epi based on microarray analysis (Karavolos et al. 2008). The PmrAB two-component signal transduction system is important for Salmonella resistance to antimicrobial peptides and virulence (Gunn 2008). Extracellular iron has been shown to activate the PmrAB regulon for prevention of iron toxicity (Wosten et al. 2000). The decreased expression of the PmrAB regulon may be due to sequestration of extracellular iron by Epi which would decrease iron binding and signaling by the PmrB sensor kinase.
7.2.8 Epi- and NE-Regulated Genes Identified by Transposon Mutagenesis
A screen of 10,000 S. Typhimurium MudJ transposon mutants identified seven fusions down-regulated and one fusion up-regulated in the presence of ~250 μM Epi (Spencer et al. 2010). The down-regulated genes included virK, mig14, iroC (see NE-enhanced growth above), accC, nrdF, yedP, and STM3081; the yhaK gene was up-regulated. Analysis of transcriptional activity using β-galactosidase assays following a 30 min exposure during mid-log phase to either 50 μM Epi or NE was confirmed in M9 minimal medium for down-regulated genes and LB medium for the up-regulated gene, yhaK. The regulation of yhaK, virK, and mig14 by 500 μM Epi and NE could be reversed in a promoter-luciferase fusion assay by addition of 500 μM phentolamine, an α-adrenergic antagonist. Both virK and mig14 are Salmonella virulence genes involved in bacterial resistance to antimicrobial peptides (Brodsky et al. 2002; Detweiler et al. 2003). Exposure to 500 μM Epi or NE significantly increased sensitivity of wild-type S. Typhimurium to the antimicrobial peptide cathelicidin LL-37 (Spencer et al. 2010). Furthermore, a significant increase in sensitivity to LL-37 was demonstrated for the virK mutant compared to wild-type S. Typhimurium in the absence of catecholamines.
7.2.9 Horizontal Gene Transfer
Exposure to 5 μM NE in LB medium significantly increased conjugation frequency of a plasmid encoding multidrug resistance from a donor S. Typhimurium strain to a recipient E. coli strain (Peterson et al. 2011). Enhanced plasmid transfer was associated with a significant up-regulation of tra gene expression involved with plasmid transfer; specifically, the transcription of traGIJRY was increased upon exposure to NE. Treatment with 500 μM phentolamine reduced the NE-enhanced conjugation frequency to baseline levels as did propranolol but this effect was delayed. Treatment with adrenergic antagonists did not reduce the baseline conjugation frequency of the S. Typhimurium donor. This study suggests that bacterial exposure to catecholamines may influence the evolution and adaptation of pathogens in the environment due to the transfer of genes that encode resistance to antibiotics and virulence factors.
7.3 Escherichia coli
7.3.1 Experimental Conditions
Investigations of E. coli exposure to catecholamines have also used various media and concentrations of catecholamines. The experimental conditions for DNA microarray analysis by Dowd involved a 1:50 dilution of an E. coli O157:H7 EDL933 overnight culture in serum-SAPI medium with or without 50 μM NE (Dowd 2007). The EDL933 culture was harvested following incubation for 5 h at 37 °C, 0.05 % CO2, 95 % humidity. Bansal et al. also utilized EDL933 to analyze biofilm gene expression using microarrays in the presence of 50 μM Epi, NE, or untreated controls (Bansal et al. 2007). The EDL933 biofilms were developed for 7 h on 10 g of glass wool in 250 ml LB, 0.2 % glucose with a starting bacterial turbidity of ~0.03. DNA microarrays were employed to transcriptionally analyze E. coli O157:H7 86-24 harvested for RNA extraction at O.D.600 = 1.0 following growth in low-glucose Dulbecco’s modified Eagle’s medium (DMEM) with and without 50 μM Epi at 37 °C with shaking at 250 rpm (Njoroge and Sperandio 2012).
7.3.2 Motility
Bacterial motility in the presence of Epi or NE is one of the most often investigated phenotypes of E. coli in response to catecholamines. Using an agarose plug chemotaxis assay, both Epi and NE were chemoattractants in a concentration-dependent migration of EDL933 towards the catecholamines (Bansal et al. 2007). Furthermore, Epi and NE increased motility 1.4-fold compared to the control culture on motility medium containing 1 % tryptone and 0.25 % NaCl. Microarray analysis of EDL933 biofilm cultures in the presence of Epi and NE by Bansal et al. demonstrated a significant increase in fliD encoding a flagellar hook-associated protein but a decrease in motB encoding a subunit for the flagellar proton motive force generator. Transcription of fliC encoding a flagellin protein was significantly increased approximately twofold in the presence of 50 μM Epi in DMEM compared to cultures without catecholamines (Rasko et al. 2008). The presence of 50 μM NE has been shown in multiple investigations to significantly enhance the motility of E. coli O157:H7 86-24 in DMEM motility assays (Sharma and Casey 2014a, b). However, an increase in the transcription of genes in the flagellar and chemotaxis operons is not always apparent, probably due to the experimental conditions used. Gene expression assays are usually performed using broth cultures whereas motility assays are typically performed using semi-solid agar medium. The differences in incubation conditions including growth rate and growth phase for broth and motility assays may account for a lack of congruence between transcriptional analysis and motility phenotype.
7.3.3 The QseBC Two-Component System
The role of regulatory proteins in catecholamine enhanced phenotypes including motility is an area of intense research. The QseC sensor kinase has been proposed to be a bacterial adrenergic receptor (Clarke et al. 2006). Multiple investigations using EHEC and UPEC isolates have demonstrated that qseC mutants have decreased motility compared to wild-type E. coli (Sperandio et al. 2003; Hughes et al. 2009; Kostakioti et al. 2009; Hadjifrangiskou et al. 2011; Guckes et al. 2013). Inactivation of the qseC gene in the presence of an active QseB response regulator results in pleiotropic effects including virulence attenuation, metabolic dysregulation and decreased motility (Kostakioti et al. 2009; Hadjifrangiskou et al. 2011). Multiple physiological pathways are perturbed in qseC mutants, including a compromised TCA cycle. Interrogation of the TCA cycle in E. coli UPEC revealed that ΔsdhB and Δmdh mutations confer virulence attenuation and decreased motility compared to the wild-type strain (Hadjifrangiskou et al. 2011); these phenotypes are similar to those of a qseC mutant, indicating that the presence of an active QseB in the absence of QseC results in decreased motility and metabolic dysregulation. In support of this hypothesis, qseB mutants of both EHEC and UPEC do not have decreased motility or virulence attenuation compared to wild-type E. coli (Hughes et al. 2009; Kostakioti et al. 2009). Furthermore, Sharma and Casey demonstrated that an E. coli O157:H7 qseBC mutant has a similar motility phenotype on DMEM motility medium compared to wild-type O157:H7 (Sharma and Casey 2014b). However, in response to 50 μM NE, Sharma and Casey demonstrated a significant increase in the motility of E. coli O157:H7 qseC and qseBC mutants (Sharma and Casey 2014a, b). These results suggest that either QseC is not a sensor for catecholamines or as has been suggested, multiple regulatory systems sense and response to Epi and NE in E. coli (Njoroge and Sperandio 2012; Karavolos et al. 2013). Due to the pleiotropic effects displayed by a qseC mutant, a clear consensus concerning the role of specific two-component systems in catecholamine sensing is lacking. For this reason, differential expression of genes regulated by the QseBC and other two-component systems in response to catecholamines will not be further discussed for E. coli. Instead, readers are referred to publications by Hughes et al. and Njoroge and Sperandio for further information (Hughes et al. 2009; Njoroge and Sperandio 2012).
7.3.4 E. coli O157:H7 Locus of Enterocyte Effacement (LEE)
The locus of enterocyte effacement (LEE) is a pathogenicity island that contains multiple operons and is present in enteropathogenic (EPEC) and various enterohemorrhagic E. coli (EHEC). As shown by quantitative RT-PCR, exposure of EHEC strain 86-24 to 50 μM Epi increased the expression of the ler gene encoding a regulator of LEE expression by ~1.5-fold (Rasko et al. 2008). Microarray analysis and quantitative RT-PCR by Dowd demonstrated that the most highly induced genes in EDL933 due to 50 μM NE exposure were espAB encoded in the LEE4 operon (Dowd 2007). The eae gene in the TIR operon was also highly expressed in the presence of NE. Using microarrays Njoroge and Sperandio indicated that exposure of 86-24 to 50 μM Epi increased the expression of LEE genes and non-LEE encoded virulence effectors (Njoroge and Sperandio 2012). Quantitative RT-PCR confirmed that expression of the LEE effector espA and the non-LEE effector nleA were increased two- and sixfold in the presence of Epi, respectively.
7.3.5 Shiga Toxin and the SOS Response
The shiga toxins Stx1 and Stx2 are encoded within lambdoid prophages integrated into the chromosome of E. coli O157:H7 strains. Similar to other phage-encoded genes, the regulation of stx1 and stx2 may be stimulated by environmental stresses that induce an SOS response. Microarray analysis by Dowd in serum-SAPI medium demonstrated that transcription of stx1, stx2, umuD, recB, and several phage-encoded gene products (endolysins and holin) were increased in EDL933 due to 50 μM NE. Induction of the SOS response is typically due to environmental stresses that induce DNA damage and the concomitant response to repair damage to nucleic acids. One possible explanation for induction of DNA damage is the enhanced bioavailability of iron due to NE with elevated intracellular iron increasing the vulnerability of bacterial DNA to oxidative damage (Touati et al. 1995). Dowd also suggested that NE could be an inducer of a positive adaptive state.
7.3.6 Iron Acquisition
Regulation of iron uptake and utilization genes is a common theme following exposure to catecholamines, and microarray analysis by both Dowd and Bansal et al. confirmed this effect (Dowd 2007; Bansal et al. 2007). Exposure of biofilms by Bansal et al. to 50 μM Epi or NE induced feoAB, fhuBCD, and additional iron regulated genes. Cultures of EDL933 grown in serum-SAPI medium in the presence of 50 μM NE increased expression of fecCD, fhuD, and feoB; other iron-regulated genes were down-regulated by NE including fepAC, entCD, and the ferric uptake regulator fur. The results of Dowd are consistent with the iron deplete conditions of serum-SAPI medium due to transferrin present in mammalian serum that sequesters iron from the bacterial cell in opposition to the property of NE to bind iron and promote the growth of E. coli.
7.3.7 Cold Shock
Investigations by both Bansal et al. and Dowd found that genes encoding cold shock proteins were induced due to exposure to 50 μM Epi and NE (Bansal et al. 2007; Dowd 2007). Bansal et al. demonstrated that the expression of the cold shock regulator genes cspGH were increased 6- to 23-fold due to the presence of Epi or NE. In addition, the cold shock genes cspE and deaD were also up-regulated due to catecholamine exposure. Dowd demonstrated that the expression of cspG and cspH increased 3.4- and 3.6-fold in the presence of NE, respectively.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree