Edward L. Murphy, Roberta L. Bruhn
Human T-Lymphotropic Virus (HTLV)
Human T-lymphotropic virus type 1 (HTLV-1), the first recognized human retrovirus, was described in 1979 by Poiesz and co-workers,1,2 who isolated retroviral particles with type C morphology budding from fresh cultured lymphocytes of a 28-year-old American man with a diagnosis of cutaneous T-cell lymphoma similar to cases seen in persons residing in Caribbean countries. Soon thereafter, Yoshida and associates3 and Watanabe and colleagues4 similarly isolated a new retrovirus from cases of adult T-cell leukemia (ATL) that was common in southwestern Japan at that time, and the two isolates were subsequently determined to be the same virus. The long-standing search for a human homologue to cancer-causing retroviruses of animals, first discovered at the beginning of the 20th century, ended at a time when most researchers had abandoned this quest and focused instead on viral transforming genes that occur as oncogenes in human tumors. HTLV type 2 (HTLV-2) was identified 2 years later from a case of variant T-cell hairy cell leukemia and has a different epidemiology and disease profile compared with HTLV-1.5 More recently, rare isolates of two new HTLV types have been found among central African hunters and designated HTLV-3 and HTLV-4 in 2005.6,7 The techniques used to isolate and characterize these viruses provided the intellectual and technical basis for the discovery of the human immunodeficiency virus (HIV) in 1983,8,9 which was shown to be the cause of acquired immunodeficiency syndrome (AIDS) in 1984.10
Within the taxa of DNA and RNA reverse-transcribing viruses, the HTLV viruses, along with bovine leukemia virus, are classified in the subfamily Retroviridae within the genus Deltaretrovirus (formerly Oncovirus).11 The oncogenic properties of these viruses and their molecular structure distinguish them from retroviruses HIV type 1 (HIV-1) and HIV type 2 (HIV-2), which are members of the genus Lentivirus. Both oncoviruses and lentiviruses are capable of prolonged asymptomatic infection.11 In vitro, however, HIV-1 and HIV-2 have cytopathic effects on human T cells, whereas HTLV-1 and HTLV-2 are capable of transforming T cells, resulting in immortalized cell lines.
HTLV-1 has been shown to be associated with ATL, as well as a unique form of progressive neurologic disease known as HTLV-associated myelopathy (HAM), also known as tropical spastic paraparesis (TSP). HTLV-2 is associated with HAM but is not known to cause leukemia or lymphoma. Although most HTLV-1 and HTLV-2 infections are asymptomatic, chronic infection with either retrovirus may cause other more unusual immunologic and infectious complications.12
Structure and Molecular Organization
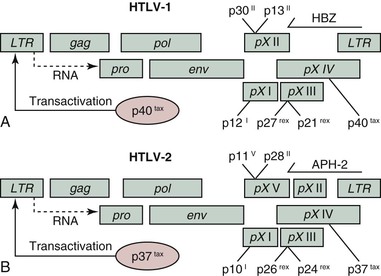
The HTLV viruses are approximately 100 nm in diameter with a thin electron-dense outer envelope and an electron-dense roughly spherical core (Fig. 170-1). Their genomic structure is illustrated in Figure 170-2. The total provirus genome consists of roughly 9000 nucleotides with two flanking identical sequences. Long terminal repeats (LTRs), at the 5′ and 3′ ends of the genome, are organized into three identical regions (U5, R, U3) that control gene expression through the requisite signals: enhancer, promoter, transcription initiation, and transcription termination and polyadenylation. Overall, there is 65% nucleotide homology between sequenced HTLV-1 and HTLV-2 isolates, 60% between HTLV-1 and HTLV-3,13 and 62% to 71% between HTLV-4 and HTLV-1, HTLV-2, and HTLV-3, respectively.14 Retroviral genes generally code for large overlapping polyproteins that are processed by a virally encoded protease and cellular proteases into functional peptide products. The HTLV viruses share with other replication-competent retroviruses the main genomic open reading frames (ORF) of gag (group-specific antigen), pro (protease), pol (polymerase), and env (envelope) encoding for structural and enzymatic proteins (Table 170-1). However, unlike other vertebrate leukemia viruses, these deltaretroviruses have an additional region called pX that contains four small ORFs: pX I, pX II, pX III, and pX IV.15 The pX ORFs III and IV encode two transcriptional regulatory proteins, TAX and REX, which are involved in regulation of virus expression. As shown in Figure 170-2, two overlapping reading frames are involved in the expression of both of these gene products translated from a doubly spliced messenger RNA (mRNA) involving the initiation codon from env and the remaining sequences from the pX region. The pX ORFs I and II code for other accessory and regulatory genes whose protein products appear to involve cell-cycle regulation.16 In addition, ORFs encoding the regulatory proteins HBZ (HTLV-I basic zipper factor), APH-2, APH-3, and APH-4 (antisense protein of HTLV) transcribe from the minus strand of the 3′ LTR in HTLV-1, HTLV-2, HTLV-3 and HTLV-4, respectively.17 The detailed roles of these proteins are still being elucidated but may involve transcriptional regulation.
TABLE 170-1
Major Structural and Regulatory Proteins of HTLV-1
| VIRAL GENE | GENE PRODUCT (PROTEIN SIZE [kDa]) | FUNCTION |
| 5′ LTR | Regulation of viral gene expression and regulation of viral expression | |
| 5′ LTR and 3′ LTR | Integration points for provirus into host genome | |
| hbz | HBZ | hbz mRNA is found in ATL tumor tissue and is responsible for lymphocyte proliferation |
| HBZ protein prevents/suppresses Tax-mediated viral transcription | ||
| gag | p15 | Nucleocapsid (NC) is a small basic protein found in the virion in association with genomic RNA |
| gag | p19 | Matrix (MA) protein is myristylated and anchored in the plasma membrane |
| gag | p24 | Capsid (CA) protein forms the major internal structural feature of the core shell of the virion |
| gag | p53 | Gag precursor |
| pro | Protease | Cleaves GAG precursor into CS, MA, NC and Pol precursor in reverse-transcriptase and integrase proteins |
| pol | Integrase | Integrates viral DNA into the host-cell genome |
| pol | Reverse transcriptase | Reverse transcriptase generates a double-stranded DNA from the single-stranded RNA genome |
| env | gp21 and gp46 | Envelope transmembrane and surface glycoproteins |
| pX | TAX (p40) | Transactivator that enhances transcription of viral promoter and alters transcription from cellular gene promoters |
| pX | REX (p27) | Regulator of unspliced and singly spliced viral mRNA transport from the nucleus to the cytoplasm |
| pX | p12I | Downregulates major histocompatibility complex class I trafficking to the surface to evade CTLs; elevates cytoplasmic calcium, increases T-cell activation, increases NFAT activation, enhances LFA-1–mediated T-cell adhesion, and activates STAT5b |
| pX | p8 | Cleavage product of p12; increases formation of intercellular conduits and viral transmission |
| pX | p30II | Interferes with TLR4 signaling, alters cell cycle and DNA repair, and promotes viral latency by interfering with tax/rex RNA export |
| p13II | Induces apoptosis; binds to and interferes with tax transactivation. |
ATL, adult T-cell leukemia; CTLs, cytotoxic T lymphocytes; LFA-1, lymphocyte function–associated antigen 1; NFAT, nuclear factor of activated T cells; TLR4, Toll-like receptor 4.
gag, pro, pol, and env
The gag gene encodes structural proteins of the nucleocapsid, capsid, and matrix, also named p15, p24, and p19 GAG proteins, respectively. The pro gene slightly overlaps gag and pol and encodes protease, which cleaves GAG and GAG/POL polypeptides into proteins of the mature virion. The pol gene encodes the enzymes reverse transcriptase, which generates a double-stranded DNA from the RNA genome, and integrase, which integrates viral DNA into the host-cell chromosomes. The polymerase region contains the largest ORF in the HTLV genome, potentially able to encode an 896-amino-acid product for HTLV-1 and a 982-amino-acid product for HTLV-2. The polymerase genes of HTLV-1 and HTLV-2 share only 56% homology based on their predicted amino-acid sequences. The env gene encodes the major components of the viral coat: the surface glycoprotein of 46-kDa (gp46) and the transmembrane glycoprotein of 21-kDa (gp2l). Although HTLV-1, HTLV-2, HTLV-3, and HTLV-4 share the same overall genetic organization, they show some diversity at the nucleotide level, exhibiting a variable degree of amino-acid homology between viral capsid and envelope proteins.
Production of the GAG proteins derives from the translation of the full-length mRNA, which yields a large precursor polypeptide that is subsequently cleaved by the virally coded protease. POL proteins are expressed as polyproteins, both GAG-PRO and GAG-PRO-POL, and production is mediated by ribosomal frameshifts between gag and pro ORFs. For the POL proteins, production depends on translation made possible when the stop codon of the gag gene is bypassed, leading to a large polypeptide including GAG- and POL-related proteins, which are subsequently cleaved into functional proteins by the viral protease. Production of the ENV surface and transmembrane proteins involves translation of a spliced message (see Fig. 170-2), which results in an envelope precursor cleaved into the subunits. The precursor proteins have characteristic molecular weights that Western blot (WB) analysis can detect immunologically.
TAX
HTLV-1 TAX is a 40-kDa protein (p40), and HTLV-2 TAX is a 37-kDa protein (p37).18,19 These proteins localize primarily to the nucleus of infected cells, although small amounts of TAX have been found in the cytoplasm. The TAX proteins are responsible for enhanced transcription of viral and cellular gene products and are essential for transformation of human T lymphocytes.20 The TAX viral regulatory protein for HTLV-1, like its counterpart TAT of HIV-1, plays an important role in promoting viral growth and disease pathogenesis. Both promote trans-acting, transcriptional activation of the LTR, but the effect of TAX appears to be mediated via expression of cellular growth factors that are abundantly activated by TAX through its trans-activation properties. This trans-activation of cellular genes by TAX not only facilitates viral replication but also has emerged as a cofactor in disease pathogenesis. The tax gene is responsible for the trans-activation of virus transcription via tax-responsive elements of a number of regulatory enhancers, such as the 21-bp enhancer, the nuclear factor-κB (NF-κB) binding site, and serum-responsive element. Such promoter interactions lead to the activation of a number of cellular genes, such as those encoding interleukin (IL)-2 and the IL-2 receptor (IL-2R), which promote cell proliferation. Additionally, TAX activates the proto-oncogenes fos and erb, as well as the gene for granulocyte-macrophage colony-stimulating factor, an array of early response genes, the human lymphotoxin gene, and parathyroid hormone–related protein gene, whereas it trans-represses the β-polymerase gene. Overproduction of interferon-γ via this pathway has been implicated in promoting the chronic inflammation that characterizes diseases such as HAM.21 The mechanism by which TAX interacts with a variety of cell regulatory elements involves nuclear regulatory elements (the NF-κB pathway); TAX also operates cytoplasmically through the induction of nuclear translocation of active transcriptional factors but not via pathways typical of other oncoviral proteins that target tumor suppressor genes.22
REX Proteins
The rex gene of HTLV-1 and HTLV-2 encodes two protein species in each virus. In HTLV-1, a 27-kDa protein (p27) and a 21-kDa protein (p21) seem to result from the use of alternative initiator methionine codons. In HTLV-2, however, a 26-kDa protein appears to be formed by phosphorylation of a serine residue in a 24-kDa protein.23 Unlike the products of the tax gene, REX does not directly regulate RNA transcription but instead appears to act chiefly at a post-transcriptional level to regulate viral gene expression. The REX (regulator of expression of virion proteins for HTLV) stabilizes viral mRNA and is essential for export of full-length gag/pol and single-spliced env mRNA from the nucleus to the cytoplasm.24 REX localizes to the nucleus and specifically to the nucleoli of infected cells.25,26 Phosphorylated REX binds with high affinity to cis-acting RNA sequences, called REX-response elements (RxRE), in the viral mRNA.27–29 This interaction facilitates the export of mRNA. REX binding may also inhibit mRNA splicing by preventing early steps in spliceosome assembly.30 As a consequence of the accumulation of REX in the cell, there is an accumulation of unspliced and single-spliced mRNA, favoring the production of structural proteins (GAG and ENV). This is accompanied by a decrease in the levels of double-spliced mRNA encoding TAX and REX. REX accumulation may also inhibit TAX, thus slowing viral transcription.31 A fine balance between TAX and REX expression and function may dictate the rate of viral replication within infected cells.
Proteins Encoded by the pX Region
pX ORF I, produced by a double-splicing mechanism similar to that of the gene products of pX ORF III and IV, codes for a hydrophobic 12-kDa protein, p12I, and pX ORF II results in the production of two nuclear proteins, p13II and p30II.16 In addition to activating nuclear factor of activated T cells,32 p12I localizes in the endoplasmic reticulum and cis-Golgi apparatus and elevates cytoplasmic calcium that is antecedent to T-cell activation and essential for establishing persistent infection.33 Other major structural and regulatory proteins of HTLV-1 are summarized in Table 170-1. The gene products of pX ORF I and II also appear to affect cell proliferation and modulate host immune responses to HTLV-1 infection.34 TAX and the ORF I and II gene products may play an integral role in the pathogenesis of HTLV-associated diseases through their effects on cyclins, which regulate cell growth. The antisense proteins HBZ in HTLV-1 and Aph in HTLV-2, HTLV-3, and HTLV-4 may also be involved in HTLV pathogenesis with effects on proviral load and transcriptional regulation. Analyses of T-cell lines transfected with mutated hbz genes showed that hbz promotes T-cell proliferation in its RNA form, whereas HBZ protein suppresses TAX-mediated viral transcription through the 5′ LTR.35 Thus far, HBZ has been shown to inhibit TAX-mediated activation of the HTLV-1 LTR, activate cellular transcription, and promote T-lymphocyte proliferation, whereas all APH proteins have demonstrated the ability to repress TAX-mediated viral transcription.17,36–39 In HTLV-2, although APH-2 is associated with higher proviral load, it has not been shown to promote cell proliferation or lymphocytosis.39,40
Biology
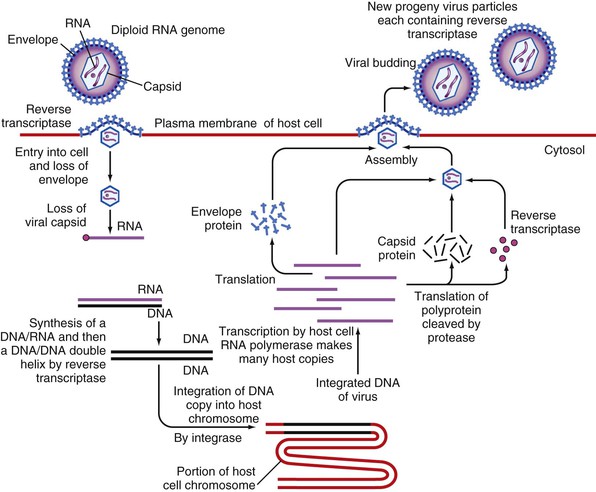
The replication strategy of the HTLVs involves a life cycle typical of all members of the Retroviridae (Fig. 170-3), whereby the RNA genome undergoes reverse transcription into a DNA provirus that integrates into the host genome. Subsequently, new virions are produced via this integrated DNA template under the regulation of viral regulatory genes. The HTLV-1 receptor was determined to be the ubiquitous glucose transporter GLUT-1,41 consistent with the infection by HTLV-1 of a wide range of cells in vitro, including endothelial cells and fibroblasts.42 Additionally, a number of animal species can be infected either experimentally (mice, rats, rabbits, and New World primate species) or naturally (Old World primates).43 The natural host range for HTLV is therefore humans and Old World nonhuman primates.
Although HTLV-1 can infect a number of different cell types in vitro, its growth and propagation in vivo are supported mainly by CD4+ cells and, to a lesser extent, CD8+ cells, as determined by real-time quantitative polymerase chain reaction (PCR) assay performed on flow cytometry–sorted T cells from patients with HAM.44 In contrast, HTLV-2 preferentially infects CD8+ cells over CD4+ cells.45
Clonal expansion of infected lymphocytes, rather than viral replication and infection of new lymphocytes, has been demonstrated as the primary mechanism of viral expansion within chronically infected humans, especially in HTLV-1.46 Limited numbers of large clones have been found in HTLV-1 carriers as well as patients with HAM/TSP,47 and the relationship between number and size of clones may be related to mode of transmission and to disease pathogenesis.48,49
Based on epidemiologic data demonstrating that HTLV-1 transmission is strongly cell associated and through in vitro studies in which cocultivation is required for efficient infection of target cells, transmission of HTLV is mediated by live cells and not via cell-free body fluids. For this reason, HTLV-1 is not an easily transmitted virus, and universal biohazard precautions are adequate for the inactivation of potentially infectious blood or bodily secretions. Procedures that remove or kill lymphocytes, such as leukoreduction or refrigerated storage of blood products and freeze/thawing of expressed breast milk, have been shown to reduce the risk for HTLV-1 transmission.50–52 Because HTLV-1 is highly cell associated, the means for viral attachment are not well characterized, but similar to other retroviruses, fusion of the virion with the cell membrane results in uncoating of the diploid RNA genome of the virus (see Fig. 170-3). Igakura and co-workers53 found that additional cell surface-adhesion proteins and cell-cell contacts/interface (virologic synapse) are important for facilitating virus transmission. Cytoskeletal reorganization in the infected cells and segregation of virus particles to the interface between the infected and uninfected cells can be observed by immunofluorescence microscopy.53–55
Once in the cell, the virally encoded RNA-dependent DNA polymerase (reverse transcriptase) complexed to the genomic RNA of the virus transcribes viral RNA into double-stranded DNA. This double-stranded viral complementary DNA (cDNA) is transported to the nucleus as a ribonucleoprotein complex that includes the p24 capsid protein, as well as integrase and reverse transcriptase. Once there, the cDNA, through a complex process mediated by the viral integrase, is inserted into the host genome. The genomic integration of HTLV-1 establishes a lifelong infection and is integral to both the virus replication cycle and amplification of provirus.
Elements in the viral LTR are essential to integration and replication, because they form the sites for covalent attachment of the provirus to cellular DNA and provide important regulatory components for transcription. Additional key regulatory elements of HTLV are tax, which activates transcription of the viral genome, and rex, which modulates the processing of the viral RNA expressing unspliced forms of the viral mRNA. When the DNA provirus is expressed (i.e., transcribed by a cellular RNA polymerase), viral genomic RNA, mRNA, and, subsequently, viral proteins are made by the cell. Under the influence of REX, which stabilizes viral mRNAs and regulates their splicing and transport, new genomic RNA is assembled at the cell membrane and packaged for release. During the budding process, the envelope incorporates some of the cell’s lipid bilayer, producing an infectious virion of about 100 nm (see Figs. 170-1 and 170-3).
Laboratory Detection
Because HTLV-1 and HTLV-2 cause chronic infections and stimulate antibody production in most infected hosts, laboratory diagnosis can rely on detection of virus by culture of lymphocytes, anti-HTLV antibody, or HTLV genetic sequence by nucleic acid testing. Each approach has strengths and limitations, and diagnosis needs to be tailored to the specific clinical or epidemiologic setting.
Virus Isolation
Direct detection of virus by culture is intensive, expensive, and time-consuming, often requiring several weeks for results, which even then may be negative in infected persons. It is therefore impractical for clinical diagnosis in most clinical situations and is reserved for research. The ability to culture retroviruses has been improved by cocultivation of patients’ T cells with human peripheral blood mononuclear cells (PBMCs) that have been stimulated in vitro with mitogens (e.g., phytohemagglutinin) and growth factors (e.g., IL-2), as well as by the removal of patients’ CD8+ suppressor cells from the coculture. The number of infected cells present in the blood of an infected individual is generally relatively low; mean proviral loads are 3.28 log10 copies per million PBMCs (range, 0.5 to 5.3) for HTLV-1 and 2.60 log10 copies per million PBMCs (range, 0.05 to 5.95) for HTLV-2.56 The ability to isolate HTLV is dependent on viral load, immune status, and stage of disease. In infants and children, the small volume of blood available for culture and low viral load make virus isolation especially challenging.
Serologic Assays and Antigen Detection
The most common test for HTLV-1 and HTLV-2 infection is detection of antibody. Varieties of techniques are used to detect antibodies to HTLV-1. Because resolution of HTLV infection has not been described, confirmed HTLV-1 antibody positivity can be interpreted as infection with HTLV provirus. Samples are first screened with one of several assays that use whole-virus lysates or recombinant HTLV-1 and HTLV-2 antigens. The most widely used assay for the detection of HTLV-1 in the United States is the enzyme immunoassay (EIA), which uses whole disrupted virus and recombinant antigens.57 This assay has performed with high sensitivity and specificity in the clinical setting but does not discriminate between HTLV-1 and HTLV-2 because of cross-reactive antibodies.58 Nevertheless, because false-positive results are common when using EIAs in low-prevalence populations such as blood donors, repeating the EIA and retesting repeatedly reactive EIAs with a different supplemental test is strongly recommended before informing the patient.
For HTLV-1 and HTLV-2 infection, a combination of a screening EIA followed by a confirmatory WB is the standard approach, although some investigators and blood banks prefer alternative strategies incorporating a particle agglutination test for screening or an immunofluorescence assay for confirmation.59–61 The inclusion of synthetic peptides in EIA and supplemental assays has improved performance.60–62
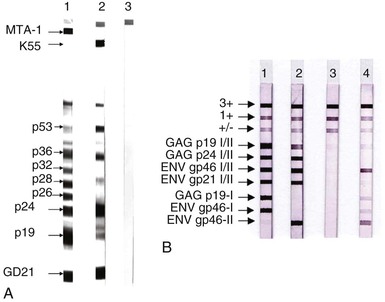
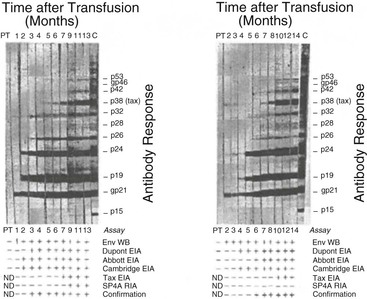
A modified WB has been developed that contains both group-specific conserved motifs from the transmembrane protein and type-specific motifs from the external glycoproteins (recombinant gp46) of HTLV-1 (MTA-1) and HTLV-2 (K55) (Fig. 170-4A). They are coated onto the strips, which allows a simultaneous confirmation and differentiation of both HTLV-1 and HTLV-2 in 98% of the cases.63 In a recent line immunoassay, antigens are purified and fixed onto a nylon membrane including two GAG bands (p19 1/2, p24 1/2) and two ENV (gp46 1/2, gp21 1/2) as non–type-specific antigens to confirm the presence of antibodies against HTLV-1/HTLV-2 (see Fig. 170-4B). The type-specific antigens GAG p19-1 and ENV gp46-1 (specific for HTLV-1) and ENV gp46-2 (specific for HTLV-2) enable differentiation of HTLV-1 and HTLV-2 infections.64 Because of the small anticipated market, neither the modified WB nor the line immunoassay has yet been licensed by the U.S. Food and Drug Administration for diagnostic use, posing regulatory problems for HTLV diagnosis by U.S. blood banks.
The criterion for WB positivity varies by manufacturer but should include the presence of reactivity to both a gag and an env gene product. In the case of env, this usually entails a recombinantly produced antigen because of the paucity of HTLV envelope antigens in whole-virus preparations. The evolution of WB band patterns in a seroconverting patient is shown in Figure 170-5.
The radioimmunoprecipitation assay, a more difficult procedure based on radiolabeled virus-infected whole cells, is more sensitive for ENV antibodies. Confirmatory testing by an immunofluorescence assay is also possible, although this assay is not suited for high-throughput operations because it is labor-intensive and subjective. Titration to allow quantification of antibody is possible by testing serial dilutions of serum with EIA or particle agglutination or immunofluorescence assays.
In tropical countries, particularly Africa, repeatedly reactive EIA-reactive samples exhibit a high frequency of indeterminate WB results. Typically, these indeterminate sera show an HTLV GAG indeterminate profile: GAG p19, p26, p28, and p30 without p24 or ENV gp21 and gp46, which may be related to endemic Plasmodium falciparum infection.65 To avoid overestimating the rate of HTLV-1/HTLV-2 seroprevalence in these regions, PCR assay may be useful.66–68
Nucleic Acid Detection
PCR assays have been developed to distinguish virus type and to quantify viral presence. In this assay, proviral DNA is amplified enzymatically and subsequently detected using a system of specific nucleotide primers and probes.69 The limit of detection is approximately one infected cell per million peripheral blood mononuclear cells.70 With the addition of a reverse-transcription step before amplification, the PCR assay has been used in the research setting to detect viral RNA in infected cells, which helps to identify actively replicating virus, although RNA has not been convincingly demonstrated in ex vivo plasma samples. Although exquisitely sensitive in the best laboratories, the PCR assay remains a research technique; it is under consideration for use as a screening assay as new technologies are evolving. PCR assay has proved valuable in enigmatic situations such as cases of suspected HTLV disease in the setting of a seroindeterminate WB result. If used with degenerate primers capable of detecting a variant of HTLV, the PCR assay can also detect the presence of a new exogenous retrovirus.65 However, the PCR assay has only occasional clinical utility in the diagnosis of HTLV infection in low-risk populations because it also is subject to false-positive results. With a low pretest probability (nonendemic area, no risk factors), individuals with positive EIA and indeterminate WB results have only a 1% to 2% probability of true HTLV infection, so the PCR assay is not recommended.71 Conversely, the PCR assay is useful in the diagnosis of neonatal HTLV infection because passively transferred maternal antibody makes antibody testing unreliable.72
The PCR technique has also been useful to facilitate epidemiologic research studies.69,73 Theoretically, virus-positive, antibody-negative individuals could be missed by antibody tests and the true prevalence of virus may be underestimated. In fact, several surveys using the PCR assay have not detected large numbers of virus-positive, antibody-negative individuals, although some instances have been reported.74–77 The recent development of quantitative real-time PCR has allowed the measurement of HTLV proviral DNA in peripheral blood mononuclear cells from infected individuals in the research setting.46,56,70,78 Unfortunately, proviral load has not yet been characterized as a useful prognostic marker and these assays are not yet available clinically.79,80
Serologic Epidemiology
Geographic Distribution
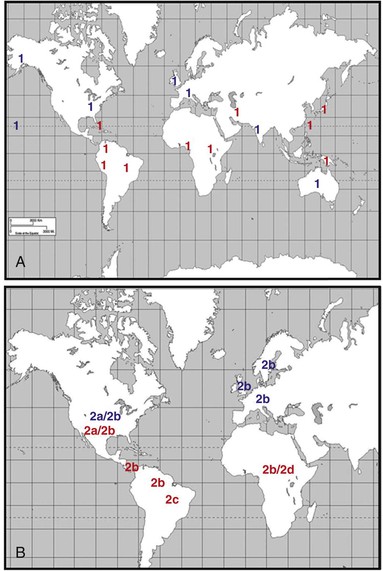
Figure 170-6 shows the different geographic distributions of HTLV-1 and HTLV-2. Although precise estimates of the global burden of HTLV are difficult, a recent review of worldwide prevalence of HTLV-1 has placed the estimate at 5 to 10 million persons.80a The largest endemic area for HTLV-1 is most probably Africa, with seroprevalence ranging from 0.5% to 5.5%. Central Africa appears to be the most highly endemic, although seroprevalence surveys have not been done in many African countries and the studies that have been done focused primarily on blood donors or pregnant women.81 Surveys from the Ivory Coast, Ghana, Nigeria, Democratic Republic of Congo, Kenya, and Tanzania document that rates of HTLV-1 seropositivity are similar to those in the Caribbean region (5% to 14%), but more and better data from sub-Saharan Africa are needed.82,83
In Asia, endemic clusters of HTLV-1 infection are present in southern Japan, the islands of the Ryukyu chain including Okinawa, and some isolated villages in the north of Japan among aboriginal Ainu populations; most of the seropositives in northern Japan are among immigrants from endemic areas in the south.84–86 Rates of infection among persons older than 40 years of age exceed 15% in these areas, and in some highly endemic areas they reach 30% to 40% in adults older than 50 years of age.81,85,86,87 Inhabitants of the People’s Republic of China and Vietnam are largely free of infection,88 whereas those living in Hong Kong and Korea have very low seroprevalence rates (0.0041% and 0.007% to 0.25% in blood donors, respectively).81,89 The rates in Taiwan are slightly higher, with 0.058% in blood donors81 and 0.82% to 1.63% in adults older than 30 years.90
In Australia, HTLV-1 seroprevalence is highly endemic in aboriginal groups of inland Australia but very low in white nonaboriginals. Seroprevalence in the Solomon Islands ranges from 1.2% to 3%, whereas the high rates (>15%) in Melanesia (primarily Papua New Guinea) are attributed to a variant HTLV-1 strain.81,91
In the Americas, a major endemic focus of HTLV-1 infection occurs in the Caribbean, where rates of seropositivity in Jamaica, Trinidad and Tobago, Barbados, Haiti, and the Dominican Republic range between 5% and 14%, most in older persons and those of African descent.92,93,94 In Jamaica, varying rates of seropositivity occur in different regions, with the highest rates (10%) of positivity observed in the lowland, high-rainfall areas.95 In Trinidad and Tobago, seropositivity (5% to 14%) is restricted almost exclusively to persons of African descent, even though individuals of Indo-Asian ethnic background have shared a common environment for more than a century.96 Seropositivity is found more frequently in persons of lower socioeconomic class and those who lack formal education.90,92,93,97,98 Men and women attending clinics for sexually transmitted infections have the highest rate of seropositivity (>15%).98
Low rates of seropositivity are present in Central America, including Panama, Honduras, Costa Rica, and Nicaragua.81 Of note is the higher rate in the non-Mestizo population compared with the Mestizo ethnic population, especially in coastal areas, in Honduras (8.1% vs. 0.5%).81 In the entirety of South America, HTLV-1 is considered endemic, but several areas are foci of high endemic rates primarily in indigenous Amerindians (Peru, Colombia, Chile, and Brazil) and, occasionally, specific groups of African origin (French Guiana, Surinam, Guyana, Colombia, and Brazil).81,99,100 Brazil is considered one of the highest areas of endemic HTLV-1 infection (especially Bahia and the northeast), whereas rates are very low in Venezuela, Paraguay, and Argentina.81,101
In North America, large-scale screening of blood donors in the United States has documented rates of HTLV-1 or HTLV-2 of up to 0.3 to 0.4 per 1000. Of the donors who test HTLV seropositive, one half to two thirds are HTLV-2 infected.103–105 In a significant proportion of HTLV-1–positive cases, the donor either has links to an endemic area or a history of risk behaviors, such as blood transfusion or multiple sexual partners.103 Smaller regional surveys and studies of military populations show patterns similar to Trinidad and Tobago; persons of African ancestry have higher rates of seropositivity. Populations migrating from Okinawa to Hawaii, from the Caribbean to the United States, and from the Caribbean to the United Kingdom are at risk for HTLV positivity, as are those who experience exposure through sexual contact or blood transfusion in viral endemic areas.97,106–110 HTLV infection is rare in Canada; when it is found it is usually in migrants from endemic areas. However, several studies have documented HTLV-1 infections in First Nations peoples in coastal British Columbia. Little information is available on HTLV prevalence in Mexico, but the few studies that have been done indicate a near absence of infection.81
In Europe, occasional infections are detected among emigrants from endemic areas. Middle East survey results have been largely negative, with the exception of Iranian Jews from northeastern Iran (Mashhad) and emigrants from that area now residing in Israel and New York.111,112 Surveys in southern India and Indonesia have identified some HTLV-1–infected individuals but not endemic foci; the Seychelles in the Indian Ocean are highly endemic for HTLV-1 (>15%).113
HTLV-2 has a more restricted distribution than HTLV-1, primarily occurring in the Americas and parts of West and Central Africa.114,115,116 A major reservoir exists in injection drug users in the United States and southern Europe, with rates ranging from 10% to 15% and higher.117,118 Amerindians residing in North, Central, and South America have varying rates of positivity for HTLV-2 (5% to 30%). Pockets of infection are present among the Seminoles in south Florida, the Pueblo and Navajo in New Mexico, and the Athapaskan in northwestern United States and Canada but not among various tribes in Alaska.119–123 In Central America, the Guaymi Indians residing in northeastern Panama near the Costa Rican border have high seropositive rates (>15%), but this does not hold true for the Guaymi living in southwest Panama. In South America, HTLV-2 is endemic in some indigenous Amazonian tribes of Peru and Brazil, as well as the Chaco Indians of Argentina, with secondary spread to other population groups in those countries.101,121 Finally, HTLV-2 has been identified in Africa, primarily among pygmy populations living in Central and Western rain forest areas.115,124
Demographic Patterns
HTLV-1 seroprevalence rates are strongly dependent on age and sex, with higher rates associated with older age and with female sex (Fig. 170-7).84,93,125 The increasing prevalence with age may be due to the accumulation of infections over the lifetime of the individuals surveyed or birth-cohort effect due to declining HTLV-1 seroprevalence over the past decades.126 The higher prevalence in females may be the result of more efficient male-to-female sexual transmission, as discussed later, or differences in sociodemographic or behavioral factors, such as duration of breast-feeding or frequency of condom use.
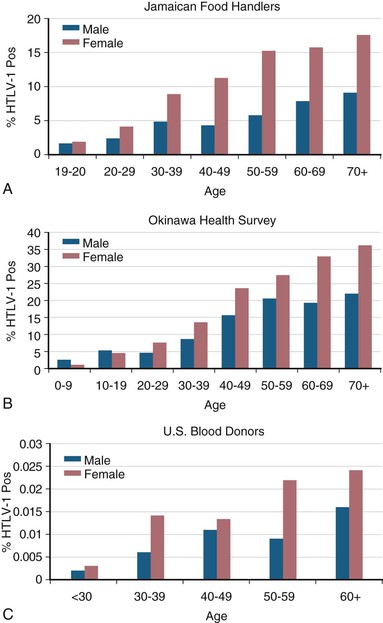
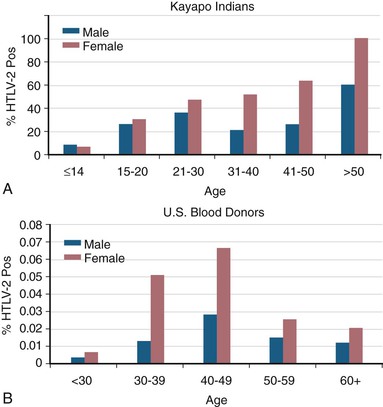
For HTLV-2, there is also a characteristic age-dependent increase in seroprevalence in endemic Amerindian populations (Fig. 170-8).127 In contrast, prevalence among U.S. blood donors shows an HTLV-2 age-specific prevalence that peaks at age 40 to 50, consistent with a birth cohort effect due to injection drug use in the 1960s and 1970s, with secondary sexual transmission.105
Molecular Epidemiology
HTLV-1
Phylogenetic analysis has been used to classify HTLV-1 into five major molecular and geographic subtypes: (1) a cosmopolitan (C) subtype isolated all over the world (endemic to the Caribbean, South America), (2) a Japanese (J) subtype, (3) a West African (WA) subtype, (4) a Central African (CA) subtype, and (5) a Melanesian (M) subtype (Papua New Guinea, Melanesia, and Australian Aboriginals).128 Other authors postulate six HTLV-1 subtypes, with a broad cosmopolitan subtype A redefined to include all Japanese isolates, four subtypes (B, D, E, and F) in Africa, and the Melanesian subtype C.129 HTLV-1 isolates from different parts of the world show a high degree of nucleotide sequence conservation, in contrast to HIV-1 and HIV-2, in which considerable genomic variability occurs. Isolates of HTLV-1 from Japan, the West Indies, the Americas, and Africa share 97% or greater homology.130–132 Even the most divergent HTLV-1, isolated in Melanesia, is 92% homologous with a prototypic Japanese isolate.133 The majority of the nucleotide differences are single point mutations that do not correlate with specific disease patterns. Studies of the LTR by restriction fragment length polymorphisms covering the major HTLV-1–endemic areas have demonstrated that variations are more linked to the geographic origin of the infected individuals than the patient’s clinical status.134–136 In addition, simian T-lymphotropic virus type 1 (STLV-1) isolates cluster phylogenically more closely with human isolates in the same geographic region than with STLV-1 isolates from different geographic regions, suggesting multiple instances of trans-species transmission.137–140 Thus, the genetic variability of HTLV-1 appears to reflect a combination of multiple trans-species transmission episodes and migration patterns of ancient populations carrying the virus.
An African origin of most HTLV-1 subtypes is supported by the occurrence of many HTLV-1 clusters in Africa67,81,141 and among persons of African descent residing in the Caribbean, but not among other migrant populations residing in the region.96 The cosmopolitan subtype was likely dispersed throughout the world during the slave trade or early maritime explorations of European countries. The high prevalence of this HTLV subtype in southwestern Japan remains unexplained and could be due either to importation by maritime trade with Portugal and Holland starting in the 16th century or to prehistoric dissemination of the cosmopolitan subtype among some ancient ethnic people of Japan.128,142,143
A rare HTLV-1 variant, subtype D, was isolated from seropositive individuals in various parts of Melanesia.144 The original isolate was obtained from an unacculturated hunter-gatherer tribe, the Hagahai, residing in the highlands of Papua New Guinea. Their only contact with the outside world had occurred within weeks of the original blood sample collection. These viruses differ by as much as 9% to 10% from the prototype Japanese strain and by as much as 4% to 6% from the viruses isolated in Australia and the Solomon Islands.144–146 HTLV-1 subtype D clusters closely with STLV-1 isolated from a stump-tailed macaque (Macaca arctoides), providing strong evidence of multiple cross-species transmissions.147 In some cases, these strains have been isolated from persons with ATL134 and HAM.148
HTLV-2
Compared with HTLV-1, the geographic origin of HTLV-2 is less certain. HTLV-2 was first documented in and is highly prevalent among intravenous drug users in the United States and Italy.118,149–153 A natural reservoir was subsequently discovered among various Amerindian populations residing in North, Central, and South America.119,121,127,154–160 The current distribution of HTLV-2 in the New World probably represents an original transmission from an endemic Amerindian to an intravenous drug user, followed by amplification of prevalence by the efficient transmission within the latter group. The presence of HTLV-2 infection in culturally and geographically distinct Indian groups in North America, Central America (Panama), and South America (Argentina, Brazil, Colombia, and Chile) led to the speculation that HTLV-2 may have originated in the New World. However, New World monkeys do not harbor STLV-2 and the discovery of HTLV-2 among Central African pygmies has discounted this hypothesis.115,124
Based on the relative divergence of nucleotide sequences of the env and LTR regions, HTLV-2 is classified into four subtypes: HTLV-2a (previously known as HTLV-2 Mo), HTLV-2b (formerly HTLV-2 NRA), HTLV-2c, and HTLV-2d.129,157,161–164 Molecular epidemiologic studies have shown that HTLV-2a is the predominant infection among intravenous drug users in urban North America and among some North American Amerindians.165 HTLV-2b is the predominant subtype in Indian groups in Panama, Colombia, and Argentina but is also found among Bakola pygmies in Cameroon and European intravenous drug users.115 HTLV-2c appears to be confined to Brazilian Indian and urban populations and is supported as a separate subtype by LTR region phylogeny but not env gene phylogenic analyses. HTLV-2d was found in an Efe pygmy in the Democratic Republic of Congo and is the closest relative to STLV-2 isolated from Bonobo chimpanzees (Pan paniscus).124
Virus isolates from these populations have revealed that HTLV-2a and HTLV-2b differ molecularly by 2% to 4%.166,167 The TAX-2 protein of HTLV-2b is 25 amino acids longer and is a more potent transactivator of the HTLV-2 LTR than the corresponding HTLV-2a protein.163 The in vivo significance of this functional difference is unknown. Conversely, HTLV-2c has LTR and env sequences related to HTLV-2a and tax sequences similar to those of HTLV-2b.157 The identical sequence of the 456-bp tax gene of HTLV-2b isolates from a Cameroonian pygmy and a Colombian Wayuu Indian support an African origin for HTLV-2.129 This, taken together with the close similarity of HTLV-2d and STLV-2, supports a scenario whereby HTLV-2 evolved into subtypes a, b, and d in Africa and was then transported to the New World whereas HTLV-2c represents a more recent variant within subtype a.
HTLV-3 and HTLV-4
Two additional HTLV types have been isolated from Central Africans living in close contact with nonhuman primates. These viruses were discovered simultaneously in 2005 by two teams working on samples derived from two inhabitants of the rain forest area in South Cameroon. Wolfe and colleagues6 described two different viral isolates from bush meat hunters, which they named HTLV-3 and HTLV-4. Calattini and colleagues7 described an HTLV-3 isolate that seems to be identical to the HTLV-3 described by Wolfe and colleagues.6 Recently, two additional HTLV-3 strains were discovered in other individuals from Cameroon.14 Reported isolates of HTLV-3 and HTLV-4 probably represent limited instances of simian to human transmission that have not become established in the human population. There has been no evidence that either new HTLV type is widely prevalent in Africa or present at all outside Africa, and current commercially available tests have not yet been adapted to detect these new HTLV types.
Routes of Transmission
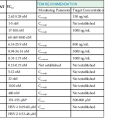
Table 170-2 summarizes the routes, mechanisms, and cofactors associated with HTLV-1 and HTLV-2 transmission. The three major routes of HTLV-1 transmission are mother-to-child transmission, sexual transmission, and parenteral transmission. Although the routes of HTLV-2 transmission have been less well studied, the available evidence suggests that they are similar to those of HTLV-1. Transmission involving casual contact does not seem to occur.
TABLE 170-2
Transmission of HTLV-1 and HTLV-2
| HTLV TYPE 1 | HTLV TYPE 2 | |
| Route of Transmission | ||
| Mother to child | ||
| Transplacental | Low efficiency | Probable, but not quantified |
| Breast milk | Efficient | Probable, but not quantified |
| Sexual | ||
| Heterosexual | Efficient | Efficient |
| Male to male | Efficient | Unknown |
| Parenteral | ||
| Blood transfusion | Very efficient | Very efficient |
| Injection drug use | Efficient | Efficient |
| Cofactors of Transmission | ||
| Elevated virus load | ||
| Mother to child | Increased | Unknown |
| Heterosexual | Increased | Increased |
| Sexually transmitted diseases | Increased | Unknown |
| Cellular versus plasma transfusion products | Increased | Increased |
| Cold storage of blood | Decreased | Decreased |
Mother-to-Child Transmission
In contrast to mother-to-child transmission of HIV-1, wherein as many as 25% of offspring of positive mothers can acquire infection without breast-feeding,168 breast-feeding is the predominant route of mother-to-child HTLV-1 and HTLV-2 transmission169,170,171,172 and occurs through ingestion of infected milk-borne lymphocytes.173 Both HTLV-1 and HTLV-2 viruses have been detected in breast milk.174–176 During the first 6 months of life, maternal antibodies are present in infants; in serial WBs, all bands often disappear before new bands appear as a result of neonatal infection. In some cases, breast-feeding had ceased up to several months before seroconversion, but a study of cells from exposed but nonseroconverting children identified none with latent HTLV-1 viral infection.74 In Japanese intervention trials,170 20% of breastfed infants seroconvert to HTLV-1, whereas only 1% to 2% of bottle-fed infants of HTLV-1–positive mothers become infected.170 In prospective studies from Jamaica, a similar rate of transmission has been documented.174
The major predictor of maternal-to-child transmission is the proviral load of the mother as measured by antibody titer and viral antigen level on short-term culture.79,177 The presence of antibody to HTLV Tax178,179 and/or Env has also been associated with transmission.174,180,181 Furthermore, the duration and timing of breast-feeding were strongly associated with the efficiency of transmission.169,171,173,182,183 In a prospective study conducted in Jamaica, among children born of HTLV-1–positive mothers in follow-up for more than 2 years, 32% of children breastfed for 12 months or longer were HTLV-1 seropositive compared with 9% of those breastfed for less than 12 months.173 These data strongly suggest that limiting the duration of breast-feeding to less than 6 months may significantly reduce mother-to-child transmission of HTLV-1. Follow-up studies indicated that seroconversion typically occurs in infants at the age of 1 to 3 years, with 2% to 5% of HTLV-1 infections resulting from mother-to-child transmission in the first few years of life.184,185 In many studies, no infants or children became newly infected after 2 years of age.172 Infection in early life may have considerable significance for subsequent risk for disease, particularly ATL.186,187
HTLV-2 transmission from mother to child also appears to occur through breast-feeding but has been less studied.188 In a small study of infected mothers, one of seven breastfed children was seropositive for HTLV-2, whereas one of 28 nonbreastfed children was seropositive.189 In another study, no HTLV-2 transmissions were found among 19 infected mothers and 20 nonbreastfed children.190 Among the Gran Chaco Indians of Argentina and the Kayapo Indians of Brazil, high rates of mother-to-child transmission have been observed (30% and 46%, respectively), suggesting the predominant role of breast-feeding in maternal-to-child transmission in these populations.158,191 Additionally, among the Guaymi Indians of Panama, there is a 1% to 2% prevalence among preadolescent children.192 This is consistent with early life infection and an excess of seropositive children when the mother is seropositive compared with the virtual absence of seropositive children when the mother is negative.184,187
Sexual Transmission
Several markers of sexual activity have been associated with transmission of HTLV-1 infection, including the number of lifetime sexual partners98,193 and ulcerative (syphilis, herpes simplex virus type 2, and chancroid) and nonulcerative (gonorrhea and Chlamydia) sexually transmitted diseases.98,194–196 Virus-positive mononuclear cells have been detected in semen, which suggests that seminal fluid is a likely vehicle for transmission.197 The presence of HTLV-1 DNA has also been found in cervicovaginal secretions from infected commercial sex workers in Peru.198,199 Although some studies have suggested that male-to-female transmission occurs more efficiently than female-to-male,200–203 this difference may be less than previously thought.204 In a 10-year follow-up study of 30 discordant heterosexual couples, the incidence of HTLV-1 infection was 0.9 per 100 person-years (95% confidence interval [CI], 0.1 to 3.3), with one male-to-female and one female-to-male transmission.204 Male-to-male sexual transmission may also occur and is supported by the higher prevalence of HTLV-1 among men who have sex with men (15%) in Trinidad compared with the general population prevalence of 2.4%.193 HIV-1 shares these routes of infection but appears to be an order of magnitude more infectious than HTLV-1.193,194 This may reflect differences in viral load or that HTLV-1 is highly cell associated, whereas HIV-1 is both cell associated and cell free.
There are also other risk factors associated with sexual transmission of HTLV-1. In a study of married couples, there was a nearly 12-fold higher risk for infection in wives of seropositive husbands older than 60 years of age, possibly because of increased viremia with age or postmenopausal changes in the vaginal epithelium.200 Higher proviral load and duration of relationship are also associated with increased transmission. In a prospective study of HTLV-infected men and their female sex partners, transmitters had higher proviral loads and had been in their relationships longer than nontransmitters.202 Another prospective study demonstrated that HTLV-1 proviral loads were two log10 lower in newly infected partners than in their positive partners who transmitted HTLV, suggesting that a small dose of sexually transmitted HTLV produces a lower proviral load setpoint.204 In addition, the presence of anti-TAX antibody has been shown to be associated with heightened transmission, possibly related to a state of virus proliferation induced by TAX and measured indirectly by anti-TAX antibody.205 Host-related factors may also play an important role in determining HTLV-1 proviral load. A study evaluating the sequence of the gp46 coding region among 13 infected patients and their partners revealed that, although the gp46 sequences were identical in each married couple, the level of HTLV-1 proviral DNA in the spouses often differed.206
Sexual transmission of HTLV-2 has been difficult to study because of the frequent coincidence of injection drug use in the study populations. However, a recent 10-year cohort study of 55 serodiscordant heterosexual couples reported two HTLV-2 transmissions, with one male-to-female and one female-to-male transmission (0.5 per 100 person-years), which was not statistically different from those seen for HTLV-1.204 In this study, all newly infected sex partners denied any history of intravenous drug use or other parenteral exposure. Previous studies have documented an association between a history of receiving money for sex, total lifetime sex partners,207,208 and length of sexual relationships with HTLV-2 seropositivity.202,203,208 In one study of female prostitutes, intravenous drug use was the major risk factor for seropositivity.209 In a serosurvey of Guaymi Indians, it was demonstrated that among women, early age at first intercourse (<13 years), higher number of lifetime sexual partners, and higher number of long-term sexual relationships were significantly associated with HTLV-2 positivity.210 Among men, intercourse with prostitutes was associated with HTLV-2 seropositivity.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree