Fig. 8.1
Structure of hepatitis C virus (HCV) genome and viral proteins. Upon infection, uncoated viral RNA is directly translated into a precursor protein through an internal ribosome entry site (IRES)-dependent manner and processed by host and viral proteases into ten viral proteins. The structural proteins forming viral particles are processed by signal peptidase (SP). Core protein is cleaved off from the polyprotein and processed by SP and the signal sequence was further processed by signal peptide peptidase (SPP). The non-structural (NS) proteins are cleaved by NS2 and NS3/4A proteases and recruit various host proteins to make a large replication complex required for viral RNA replication. Replicated viral RNA forms nucleocapsid with core protein and generates HCV particles bearing E1 and E2 envelope glycoproteins
8.2 Core Protein Plays Crucial Roles in Pathogenesis of HCV
It is well-known that HCV mainly infects hepatocytes and induces steatosis, cirrhosis and hepatocellular carcinoma (Maasoumy and Wedemeyer 2012). Chronic HCV infection is often associated with at least one extrahepatic manifestation (EHM), including mixed cryoglobulinemia, non-Hodgkin lymphoma, lichen planus, thyroiditis, diabetes mellitus, Sjögren syndrome, and arthritis (Galossi et al. 2007). Although precise molecular mechanisms of HCV-induced pathogenesis remain unknown (Li et al. 2011), HCV core transgenic (CoreTg) mice developed insulin resistance, steatosis, and hepatocellular carcinoma (Shintani et al. 2004; Moriya et al. 1998), suggesting that HCV core protein plays a crucial role in liver diseases and EHM (Fig. 8.2).
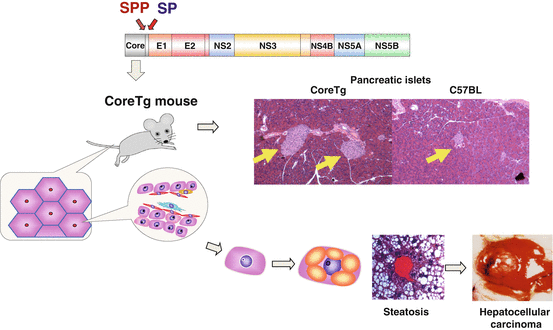

Fig. 8.2
Core protein has major roles in hepatitis C virus (HCV)-induced pathogenesis. HCV core transgenic (CoreTg) mice, which express core protein (genotype 1b) under the hepatitis B virus X promoter, have been developed. The mice show insulin resistance (2 months old~), steatosis (~3 months old), and hepatocellular carcinoma (HCC; 10–16 months old). Yellow arrows indicate pancreatic islets in CoreTg and control (C57BL) mice. CoreTg mice exhibit enlarged pancreatic islets, which is the feature of insulin resistance
8.3 HCV Core Protein Interacts with PA28γ in the Nucleus
We have shown that HCV core protein is degraded in the nucleus by the proteasome through the interaction with proteasomal activator 28γ (PA28γ) in ubiquitin/adenosine triphosphate (ATP)-independent manner (Fig. 8.3a, Moriishi et al. 2003; Suzuki et al. 2009). To understand the roles of interaction between HCV core protein and PA28γ on the pathogenesis in mice, we crossed CoreTg mice with PA28γ−/− mice and established CoreTg/PA28γ−/− mice. Although core protein localizes in the cytoplasm in CoreTg mice, the majority of core protein was detected in the nucleus in CoreTg/PA28γ−/− mice (Moriishi et al. 2007). Surprisingly, the insulin resistance, steatosis, and hepatoccellular carcinoma developed in CoreTg mice were not observed in CoreTg/PA28γ−/− mice, despite accumulation of HCV core protein in the nucleus (Moriishi et al. 2007; Miyamoto et al. 2007). Sterol regulatory element binding protein 1c (SREBP-1c), which positively regulates the production of saturated and monounsaturated fatty acids and triglycerides, is enhanced in the liver of CoreTg mice (Fig. 8.3b), leading to accumulation of lipid droplets in the liver. On the other hand, CoreTg/PA28γ−/− mice exhibit no activation of SREBP-1c. These data suggest that degradation of HCV core protein in the nucleus through the PA28γ-dependent proteasome plays a crucial role in the pathogenesis observed in CoreTg mice (Fig. 8.3a) (Mori et al. 2008).
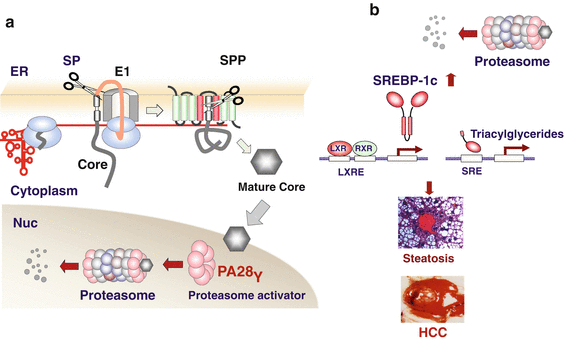

Fig. 8.3
Proteasomal activator 28γ (PA28γ) involved in core-induced pathogenesis. (a) PA28γ had been identified as a core-interacting protein by yeast two-hybrid screening. PA28γ degrades hepatitis C virus core protein in the nucleus in a ubiquitin-independent proteasome-dependent manner. (b) From our core transgenic (CoreTG)/PA28γ data, we hypothesize the following. Core protein degraded by PA28γ enhanced transcriptional activity of liver X receptor (LXR)/retinoid X receptor (RXR) complex by unknown mechanisms. The activation of LXR/RXR leads to expression of sterol regulatory element binding protein 1c (SREBP-1c), a master regulator of triacylglycerides and fatty acids production. Eventually, CoreTg mice develop steatosis and hepatocellular carcinoma (HCC)
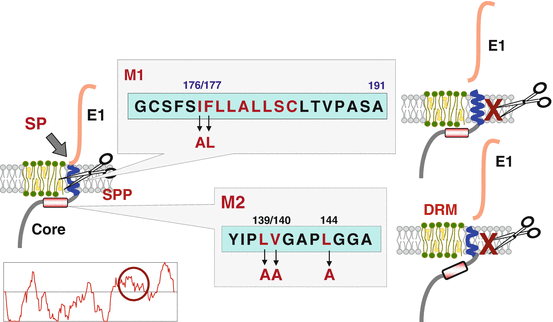
8.4 Maturation of Core Protein by Signal Peptide Peptidase
The HCV core protein is cleaved from a precursor polyprotein by a signal peptidase (SP) at amino acid position 191 to liberate it from an envelope E1 protein and then is further processed by a signal peptide peptidase (SPP) (Hüssy et al. 1996; McLauchlan et al. 2002). However, the biological significance of the intra-membrane processing of the HCV core protein by SPP remains largely unknown. The C-terminus of the HCV core protein cleaved by SPP was identified to be Phe177 by mass spectrometry. SPP cleaves the helix-breaking site of the signal peptide and mutation of Leu176 to Ala and Phe177 to Leu in HCV core protein (M1) was deduced to acquire the intact α-helix structure in the signal sequence (Fig. 8.4, Okamoto et al. 2004). As we expected, M1 was resistant to SPP cleavage. The hydrophobic amino acid residues located in the upstream of the SPP cleavage site of HCV core protein were also necessary for SPP cleavage and mutation of Ile139, Val140, and Ile144 to Ala in HCV core protein (M2) prevented from SPP cleavage. Processing by SPP is required for localization of HCV core protein on the detergent-resistant membrane (DRM) and the recombinant HCV possessing M1 or M2 mutation in core protein abrogated production of infectious particles into the culture supernatants (Okamoto et al. 2008), suggesting that intra-membrane processing of HCV core protein by SPP is required for the localization of the HCV core protein in the DRM and the viral propagation.