Kenneth McIntosh, Stanley Perlman
Coronaviruses, Including Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS)
The family Coronaviridae, within the order Nidovirales, contains two subfamilies, the Coronavirinae and the Torovirinae. Coronaviruses (CoVs) are a large group of viruses infecting mammals and birds and producing a wide variety of diseases. They have been divided into four genera, two of which contain viruses infecting humans (see later). All human coronaviruses (HCoVs) are primarily respiratory pathogens. During the winter of 2002 to 2003, an alarming new disease appeared, severe acute respiratory syndrome (SARS), which was quickly attributed to a new CoV, the SARS-CoV. The outbreak originated in southern People’s Republic of China, with evidence that the virus was first derived from bats and was transmitted to man through an intermediate host, probably the palm civet (Paguma larvata) or raccoon dog (Nyctereutes procyonoides).1–3 The SARS epidemic was controlled through a massive effort at case identification and containment, and the last known case occurred in mid-2004. In retrospect, the emergence of SARS is consistent with what is known about CoVs as a group: They are important pathogens in animals causing a wide variety of diseases through a wide variety of pathogenic mechanisms, and they have been noted to mutate frequently and infect new species.4,5
More recently, a related but different CoV producing severe respiratory disease has emerged, the Middle East respiratory syndrome coronavirus (MERS-CoV). MERS-CoV was grown in June 2012, from a sputum sample obtained from a man in Saudi Arabia who died of overwhelming pneumonia. The virus was quickly identified as a new CoV most closely related to several bat CoVs.6 This report was followed by a number of other reports identifying a total of 537 infected individuals, all of whom had acute respiratory symptoms, severe in most, and fatal in 145 (as of May 11, 2014).7 Human-to-human transmission has been documented, especially in hospital settings,7a but appears to be inefficient.
History
Respiratory Coronaviruses and Severe Acute Respiratory Syndrome
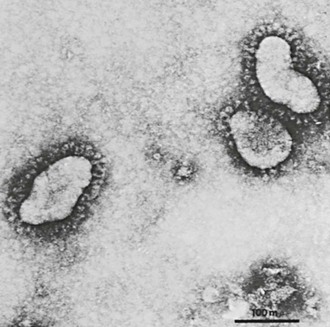
In 1965, Tyrrell and Bynoe8 cultured a virus obtained from the respiratory tract of a boy with a common cold by passage in human embryonic tracheal organ cultures. The media from these cultures consistently produced colds in volunteers. The agent was ether sensitive but not related to any known human virus. Subsequently, electron microscopy of fluids from infected organ cultures revealed particles that resembled infectious bronchitis virus of chickens.9 At about the same time, Hamre and Procknow10 recovered a cytopathic agent in tissue culture from medical students with colds. The prototype virus was named 229E and was found on electron microscopy to have a similar or identical morphology (Fig. 157-1).9
Using techniques similar to those used by Tyrrell and Bynoe, McIntosh and colleagues11 reported the recovery of several infectious bronchitis-like agents from the human respiratory tract, the prototype of which was named OC43 (OC for organ culture). At much the same time, mouse hepatitis virus and transmissible gastroenteritis virus of swine were shown to have the same morphology on electron microscopy.12,13 Shortly thereafter, the name coronavirus (the prefix corona denoting the crownlike appearance of the surface projections) was chosen to signify this new genus.14
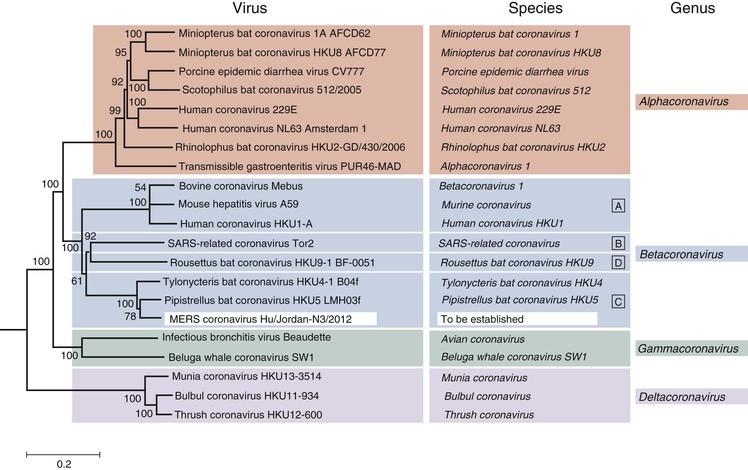
The number of animal CoVs quickly grew, including viruses causing diseases in rats, mice, chickens, turkeys, various other bird species, cattle, several wild ruminants, beluga whales, dogs, cats, rabbits, and pigs, with manifestations in the respiratory and gastrointestinal tracts, central nervous system (CNS), liver, reproductive tract, and others. Through sequencing and antigenicity studies, the animal and human CoVs (HCoVs) initially were divided into three groups: group 1, which contained HCoV-229E, as well as numerous animal viruses; group 2, which contained HCoV-OC43 plus the closely related animal viruses, bovine CoV (BCoV) and mouse hepatitis virus; and group 3, which included only avian viruses related to infectious bronchitis virus (Fig. 157-2). Current taxonomy divides the subfamily Coronavirinae into four genera: Alphacoronavirus (which includes viruses previously in group 1); Betacoronavirus (which includes viruses previously in group 2, including SARS-CoV and the MERS-CoV); Gammacoronavirus (which includes viruses previously in group 3); and Deltacoronavirus (which includes several newly described avian viruses).
SARS was first identified in Guangdong Province of the People’s Republic of China in November 2002 and spread from there to Hong Kong and then throughout the world.15 A CoV was independently and almost simultaneously isolated from SARS patients by several laboratories and found by sequencing to be only distantly related to previously characterized CoVs.16–20 The SARS outbreak stimulated a rapid and intense public health response coordinated by the World Health Organization, and by July 2003, transmission had ceased throughout the world. Despite this effort, however, 8096 probable cases had occurred in 29 countries, with 774 deaths.15
With the identification of the SARS-CoV, the HCoV field became much more active. Sensitive molecular methods were developed to detect RNA from viruses identical or closely related to HCoV-229E and HCoV-OC43 in the respiratory tract, and two new species were discovered: NL63, an alphacoronavirus, and HKU1, a betacoronavirus.21–23 HCoV-NL63 was found independently by three groups, two in the Netherlands and, somewhat later, the third in New Haven, Connecticut.24 In all three cases, positive samples were from infants and children with respiratory disease. Notably, HCoV-NL63 and HCoV-229E were estimated to originate from a common precursor and diverge approximately 1000 years ago.25 HCoV-HKU1 was found in Hong Kong in an adult with respiratory disease. These two new HCoV strains subsequently have been found worldwide and appear to have pathogenicity similar to that of HCoV-229E and HCoV-OC43, with the possible exception that NL63 is more frequently found in children with croup.26,27
The MERS-CoV was found when a man was admitted in June 2012 to a hospital in Jeddah, Saudi Arabia with overwhelming acute pneumonia with renal failure, and a sample of sputum grew a cytopathic virus that, on sequencing, proved to be a CoV, classified as a Betacoronavirus, and most closely related to two bat CoVs, HKU-4 and HKU-5.6 Over the next 23 months 536 additional cases (145 fatal) were found, all but a few of them sporadic or hospital-based and in individuals living or traveling in the Middle East.
In the remainder of this chapter, the group of respiratory HCoVs first discovered in the 1960s and containing HCoVs 229E, OC43, NL63, and HKU-1 are referred to as community-acquired HCoVs to distinguish them from the SARS-CoV and the MERS-CoV.
Gastrointestinal Coronaviruses and Toroviruses
In view of the prominence of CoVs in animal enteric diseases, there have been extensive efforts to identify enteric HCoVs. There are numerous reports of CoV-like particles (CoVLPs) found by electron microscopy in human fecal matter, but these particles have been difficult to characterize further. More recent efforts to detect CoV RNA in feces using polymerase chain reaction (PCR) and primers for respiratory HCoVs have had limited success and have failed to associate CoVs with gastrointestinal disease.28,29
Toroviruses were, like CoVs, first described in animals. They were first detected in the feces of cattle (Breda virus) and horses (Berne virus).30,31 Shortly thereafter, Beards and colleagues32 examined human fecal material and reported finding particles with a similar appearance that aggregated in the presence of antiserum to the bovine and equine viruses. The human toroviruses, like the bovine toroviruses, do not grow in tissue culture, and thus almost all existing information about them is based on electron micrographic data. Unlike animal toroviruses,33,34 PCR-amplified torovirus RNA sequences have not been found in human stool samples. A report of genome sequences amplified from particles purified from human stool35 was subsequently retracted and considered to reflect laboratory contamination from porcine strains.36 At this time, there are no reports definitively showing the existence of human toroviruses.
Description of the Pathogens
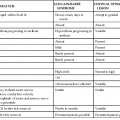
The CoV nucleic acid is RNA, approximately 30 kilobases in length, of positive sense, single stranded, polyadenylated, and infectious. The RNA, the largest known viral RNA (Fig. 157-3), codes for (in order from the 5′ end) a large polyprotein that is cleaved by virus-encoded proteases to form several nonstructural proteins, including an RNA-dependent RNA polymerase, methyltransferases, and a helicase, followed by either four or five structural proteins intermingled with a variable number of nonstructural and minor structural proteins.37 The first of the major structural proteins is a surface hemagglutinin-esterase protein, present on HCoVs OC43 and HKU1 and some animal betacoronaviruses, that may play some role in the attachment or release of the particle, or both, at the cell surface. This gene contains sequences similar to the hemagglutinin of influenza C virus, likely evidence of an interfamily recombinational event that occurred many years ago. The next gene encodes the surface glycoprotein that forms the petal-shaped surface projections and is responsible for attachment and the stimulation of neutralizing antibody. This is followed by a small envelope protein, a membrane glycoprotein, and a nucleocapsid protein that is complexed with the RNA. There are several other open reading frames whose coding functions are not clear. The strategy of replication of CoVs is similar to that of other nidoviruses, in that all messenger RNAs form a nested set with common polyadenylated 3′ ends, with only the unique portion of the 5′ end being translated.4 As in other RNA viruses, mutations are common in nature, although the mutation rate is much lower, approximately 2 × 10−6 per site per replication cycle.38 Unlike other RNA viruses, CoVs encode a 3′-5′ exonuclease that has proofreading activities, playing a critical role in maintaining replication fidelity in cell cultures and in animals.39 CoVs are also capable of genetic recombination if two viruses infect the same cell at the same time.
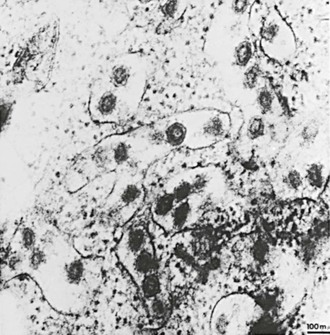
All CoVs develop exclusively in the cytoplasm of infected cells (Fig. 157-4). They bud into cytoplasmic vesicles from membranes of the pre-Golgi endoplasmic reticulum. These virus-filled vesicles are then extruded by the exocytic secretory pathway.40 The resultant virus particles have a diameter of 70 to 80 nm on thin-section electron microscopy and 60 to 220 nm on negative staining. They are pleomorphic, with widely spaced, petal-shaped projections 20 nm long (see Fig. 157-1).
The cellular receptor for 229E and most other alphacoronaviruses is aminopeptidase N (APN).41 Interestingly, NL63, the other known human alphacoronavirus, uses as its cellular receptor angiotensin-converting enzyme II (ACE2),42 the same receptor as is used by the SARS-CoV.43 Mouse hepatitis virus, a betacoronavirus related to strain OC43, uses as its receptor a member of the carcinoembryonic antigen family.44 HCoV-OC43 and BCoV, which is closely related to HCoV-OC43, bind to 9-O-acetylated neuraminic acid as part of the entry process.45 The host cell receptor for MERS-CoV is dipeptidyl peptidase 4 (DPP-4), which, like ACE2 and APN, is an ectopeptidase that is abundantly expressed in the respiratory tract.46
All the community-acquired respiratory HCoVs grow only with difficulty in tissue culture. Despite this, both 229E and NL63 were discovered because they produced a detectable cytopathic effect, the first in human embryonic kidney10 and the second in LLC-MK2 cells.21 Both the SARS-CoV and the MERS-CoV were initially isolated and grew readily in Vero cells.6,17 HCoVs OC43 and HKU-1 have been grown in tissue culture after laboratory adaptation or in primary human airway epithelial cells.47–49 Detection of all these viruses in clinical specimens is most conveniently and sensitively achieved using PCR.
Likewise, the enteric CoVs have been difficult to cultivate in vitro. All but a few strains have been detected only by electron microscopy of human fecal material.50–52,53,54 Some strains have been characterized by immune electron microscopy and found to be related to HCoV-OC43.55 Two strains obtained from an outbreak of necrotizing enterocolitis in Texas and passaged in intestinal organ cultures were reported to contain four or five proteins with apparent molecular weights similar to those of other CoVs but not related antigenically to known human strains or mouse hepatitis virus A59.56 The evidence favors the view that these isolates, as well as particles antigenically related to HCoV-OC43, are members of the family Coronaviridae, although their association with human disease is not yet proven. Surveys of children with diarrhea using PCR imply that diarrhea associated with the four well-described HCoV strains is unusual.28,29
Epidemiology
Community-Acquired Respiratory Coronaviruses
Evidence of community-acquired respiratory CoV infections has been found wherever in the world it has been sought. In temperate climates, respiratory CoV infections occur more often in the winter and spring than in the summer and fall. The contribution of CoV infections to the total number of upper respiratory illnesses may be as high as 35% during times of peak viral activity. Overall, the proportion of adult colds produced by CoVs may be reasonably estimated at 15%.
Early studies of HCoV-OC43 and 229E in the United States demonstrated periodicity, with large epidemics occurring at 2- to 3-year intervals.57 Strain HCoV-229E tended to be epidemic throughout the United States, whereas strain HCoV-OC43 appeared in localized outbreaks. Similar studies of NL63 and HKU1 have not been done, but it seems from the available data that they also vary widely in incidence from year to year and place to place. Reinfection is common and may be due to the rapid diminution of antibody levels after infection.58 Infection occurs at all ages but is most common in children. The ratio of symptomatic to total infections varies between 50% and 90%, depending on the age of the population studied, the method of virus detection, and the definition of “infection.”59,60 Among adult volunteers 72% of those infected with HCoV-229E developed colds.61
Middle East Respiratory Syndrome Coronavirus
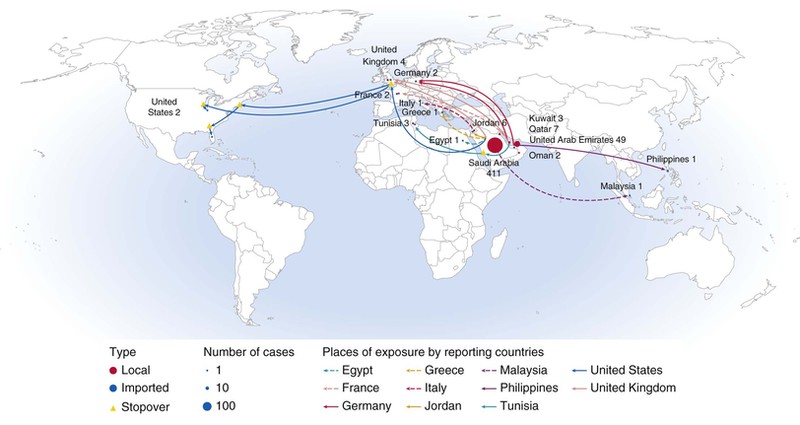
The first report of a new CoV causing severe pneumonia appeared in ProMed Mail on September 20, 2012. A man from Jeddah, Saudi Arabia had developed pneumonia in June and died of respiratory and renal failure, and a virus was grown from a sputum sample that was subsequently sequenced and found to be a betacoronavirus thought at the time to be most closely related to bat CoVs HKU4 and HKU5.6 Between then and May 2014, a total of 537 cases occurred, all infected by this virus, now termed the Middle East respiratory syndrome coronavirus (MERS-CoV). One hundred forty-five of the cases were fatal.7 More than 400 of these have been acquired and diagnosed in the Kingdom of Saudi Arabia, with most of the remainder in the United Arab Emirates, Qatar, Jordan, and Kuwait. Cases originating in the Arabian peninsula have also occurred in travelers to Egypt, Tunisia, Germany, Italy, Great Britain, Malaysia, the Philippines, and the United States, with a few secondary cases sometimes occurring in those locations through close family or hospital spread. In the United States,these include two unrelated MERS cases, and a third case related to one of these.61a While infections with severe respiratory involvement have occurred at all ages, the elderly and those with underlying conditions (diabetes, renal disease, immunosuppression) have been most often severely or fatally affected. The WHO and the CDC have published a case definition, as well as surveillance instructions to aid in epidemiologic control of the MERS-CoV.62,63
The putative bat origin of this virus was strengthened by the finding that the virus grew readily in primary bat tissue culture.64 Nevertheless, and while bats sampled in the Middle East, Africa, and Europe were found to carry viruses closely related to the MERS-CoV,65 epidemiologic studies suggested there was likely to be at least one intermediate host. Serologic and virologic studies indicated that camels in the Middle East and Africa were frequently infected by viruses very similar to some of those found in human MERS cases.65a Acquisition of MERS from camels appears likely, although the proportion of camel-acquired cases (versus those acquired from person-to-person contact or through another animal intermediate) is not clear. Case clusters indicate that person-to-person hospital spread is more common than spread within families, and that casual-contact spread is unusual.7a
Severe Acute Respiratory Syndrome Coronavirus
The SARS epidemic began in Guangdong Province in the People’s Republic of China in mid-November 2002.66 It came to worldwide attention in March 2003 when cases of severe, acute pneumonia were reported to the World Health Organization from Hong Kong, Hanoi, and Singapore. Disease spread in hospitals to health care workers, visitors, and patients, among family members, and, on occasion, in hotels, apartment complexes, markets, and airplanes. Worldwide spread was rapid but focal (Fig. 157-5). The largest numbers of cases were reported from the People’s Republic of China, Hong Kong, Taiwan, Singapore, and Toronto, Canada. The overall case-fatality rates in these locations ranged from 7% to 17%, but persons with underlying medical conditions and those older than 65 years of age had mortality rates as high as 50%. There was no mortality in children or young adults younger than the age of 24 years.
In response to the global spread and associated severe disease, the World Health Organization coordinated a rapid and effective control program that included isolation of cases, careful attention to contact, droplet and airborne infection control procedures, quarantine of exposed persons in some settings, and efforts to control spread between countries through travel advisories and travel alerts. Presumably as a result of these efforts, global transmission ceased by July 2003.
A few subsequent cases of SARS were detected, but all were either a result of laboratory spread or individual cases related to presumed contact with civet cats or other intermediate hosts. The last known case occurred in mid-2004.
Spread of SARS to humans is thought to have occurred primarily through droplet or contact transmission, with a possible role for fomites. In most instances, an individual case transmitted to very few others, although several well-documented instances of small-particle airborne transmission occurred, resulting in super-spreading events.67 Spread in hospital settings appeared to be surprisingly efficient, but it could be effectively suppressed with the enforcement of droplet and contact precautions.68 Containment measures were efficacious, in part, because patients were most contagious only after lower respiratory disease developed.69 The chain of spread was finally broken in the People’s Republic of China, the last country to experience endemic spread, in June 2003.
It now seems almost certain that the human epidemic began with the spread of a closely related bat virus first to palm civets or other animals sold in live wild game markets and then to humans in Guangdong Province in the People’s Republic of China, and that the virus adapted itself through mutation and possibly recombination, until it transmitted readily among humans.70–72 The virus that spread worldwide came largely from a single infected individual who traveled from Guangdong Province to Hong Kong and infected a large number of individuals before himself succumbing to the disease. In contrast, the virus that was epidemic in the People’s Republic of China was more variable.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree