(1)
ClinSciences, 10 avenue Victor Hugo, 21000 Dijon, France
Abstract
Peroxisome proliferator-activated receptors (PPARs), with three existing isoforms (PPAR α, PPAR β/δ, and PPAR γ), belong to the nuclear receptors subfamily. The PPAR machinery involves the binding of natural (fatty acids and prostaglandins), synthetic (mainly fibrates for PPAR α and thiazolidinediones for PPAR γ and new dual and pan-PPAR agonists), and selective PPAR modulators, coactivators, and corepressors with enzymatic activities and various metabolic transformations turning PPARs toward activation or to degradation. Among PPAR activators in clinical development for type 1 or type 2 diabetes and dyslipidemia, some have been discontinued for safety reasons or for no additional benefit over existing drugs. Knowing the pleiotropic effects of PPAR agonists, search for new natural PPAR ligands and clinical development of new target indications represent the main future of this class of drugs.
Introduction
Peroxisome proliferator-activated receptors (PPARs), a family of cell receptors, are closely involved in glucose and lipoprotein metabolism. As discussed elsewhere in this book, PPAR alpha (PPAR α) agonists, such as fenofibrate, have shown clinical benefit for cardiovascular disease in some subgroups and for the macrovascular and microvascular complications of type 2 diabetes mellitus. This chapter reviews the basic science of the PPAR system and summarizes some clinical PPAR modulating drug trials, with an emphasis on diabetes, lipoproteins, and vascular disease. The chapter is divided into five sections:
1.
PPAR gene and gene variants, proteins, and natural ligands
2.
Synthetic ligands: from PPAR activators to PPAR agonists
3.
The PPAR machinery with subsections on:
Coactivators and corepressors
Metabolic modification (phosphorylation, ubiquitination, sumoylation, and acetylation)
Partial agonists or SPPARMs
4.
Effect of PPAR agonists in diabetes:
Pharmacology, in particular, in the pancreas
Effects in type 1 diabetes
Effects in type 2 diabetes and/or dyslipidemia with products reaching clinical development
5.
Conclusions and perspectives
PPAR Gene and Gene Variants, Proteins, and Natural Ligands
Peroxisome proliferator-activated receptors (PPARs) belong to a subfamily of the nuclear receptors which includes the retinoic acid receptors, the thyroid hormone receptors, and the RevErbA-related orphan receptors [1]. The PPAR subfamily contains three isoforms, namely, PPAR α (PPARA, NR1C1), PPAR β/δ (NR1C2 identified here as PPAR δ), and PPAR γ (PPARG, NR1C3, PPAR γ1, and PPAR γ2 sub-isoforms), that are encoded by different genes on different chromosomes.
In humans, PPAR α is mapped on chromosome 22 on the regions 22q12-q13.1; 22q13.31 with a linkage group of six genes and genetic markers [2]. The human PPAR γ gene is located on chromosome 3 at position 3p25, close to the retinoic acid receptor beta (RAR β) and the thyroid hormone receptor beta genes [3–5]. Two different human PPAR γ transcripts are expressed in hematopoietic cells: a 1.85-kb transcript, which corresponds to the full-length mRNA (PPAR γ1), and a shorter 0.65-kb transcript (PPAR γ2) [5]. PPAR γ2 is mostly expressed in adipose tissue where the PPAR γ2/PPAR γ1 ratio of messenger RNA is directly correlated with body mass index and where a low-calorie diet downregulates PPAR γ2 messenger RNA in subcutaneous fat [6]. Several variants in the PPAR γ gene have been identified, with the Pro12Ala variant having been the most extensively examined in epidemiologic studies. A strong association between PPAR γ 12Ala polymorphism and a reduction in type 2 diabetes risk (odds ratio: 0.86, 95 % CI: 0.81–0.90) was described in an updated meta-analysis of 60 studies involving 32,849 subjects with type 2 diabetes mellitus (T2DM) and 47,456 control subjects evaluated by the Human Genome Epidemiology Network [7].
The human PPAR δ, which was cloned from a human osteosarcoma cell library, is located on chromosome 6 at position 6p21.1-p21.2 [8]. In the mouse, where the first PPAR, PPAR α, was identified in 1990 by Issemann and Green [9], PPAR α is found on chromosome 15, PPAR γ is located on chromosome 6 at position E3-F1, while PPAR δ is found on chromosome 17 [10]. In both human and mouse, the PPAR transcript is encoded by six exons (one in the A/B domain, two in the C domain, one for the hinge region, and two for the ligand binding domain).
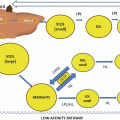
PPAR isoforms share a common domain structure as shown in the schematic view in Fig. 19.1. Five domains designated A/B, C, D, E, and F are distinguishable, and each has a different function. The N-terminal A/B domain contains at least one constitutionally active transactivation region (AF-1) and several autonomous transactivation domains (AD) [1]. The specificity of gene transcription is granted by the isoform-specific sequence of the A/B domain of the receptor [11]. Chimeric proteins generated by fusion with the A/B domains of other receptor proteins attenuate the specificity of target gene activation [11]. The DNA binding domain (DBD, C domain) is the most conserved region, which contains a short motif responsible for DNA binding specificity (P-box) on sequences called peroxisome proliferator response elements (PPREs), typically containing the AGGTCA motif.
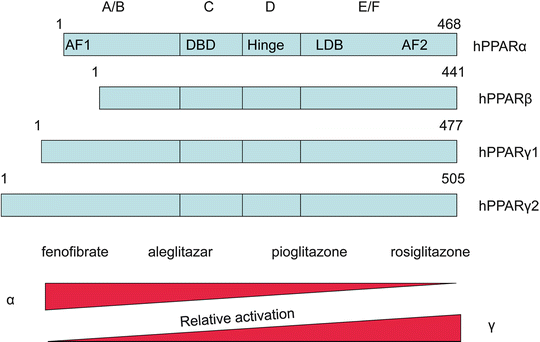

Fig. 19.1
Structure of PPARs. In the upper panel, the structure of PPARs with their four domains: 1 is the NH2 terminal and 468 the COOH terminal for PPAR α. The bottom panel illustrates the relative activation for PPAR α and PPAR γ for major agonists with fenofibrate and rosiglitazone as behaving as specific activators and aleglitazar or pioglitazone with mixed effects
The D domain, called a hinge, permits the change in shape of PPARs. The C-terminal E/F domain contains the ligand binding domain (LBD) and the AF-2 region for binding coactivators and corepressors. When activated by ligands, PPARs heterodimerize with another nuclear receptor, the retinoid X receptor, and alter the transcription of target genes after binding to specific PPREs on target genes.
Natural ligands for PPARs are long chain fatty acids, saturated or not, and eicosanoids: 8-HETE (hydroxyeicosatetraenoic acid) and to some extent leukotriene B4 (LTB4) for PPAR α; 9- and 13-HODE (hydroxyoctadecadienoic acid), two 15 lipoxygenase metabolites of linoleic acid, and 15-deoxy PGJ2 for PPAR γ; and prostacyclin (PGI2) for PPAR δ [12–14]. However, tissue concentrations are probably too low for them being the active ligands [15]. A new candidate endogenous ligand for PPAR α in the liver is a glycerophosphocholine esterified with palmitic and oleic acids 16:0/18:1-GPC or POPC (1-palmitoyl,2-oleoyl-sn-glycero-3-phosphocholine hydroxyeico-satetraenoic acid), which was identified in the liver of mice by tandem mass spectrometry [16]. This phosphatidylcholine is displaced from PPAR α by the synthetic agonist Wy14643. Its portal infusion induces dependent gene expression of carnitine palmitoyltransferase I (CPT1) in wild-type mice, but not in PPAR α deficient mice. Recently, two other phosphatidylcholines, DLPC and DUPC (1,2-dilauroyl-sn-glycero-3-phospho-choline and 1,2-(cis-cis-9,12-octadecadienoyl)-sn-glycero-3-phosphatidylcholine, respectively), have been shown to improve glucose control in two mouse models of insulin resistance [17]; however, they did not affect rosiglitazone binding to PPAR γ, and their effects are linked to stimulation of another nuclear receptor liver receptor homologue (LRH)-1.
Synthetic Ligands: From PPAR Activators to PPAR Agonists
PPAR α was first cloned from a mouse liver cDNA library at ICI, the pharmaceutical company which developed clofibrate, the first fibrate [9], and subsequently in humans [2, 18]. Fibrates, which were in clinical use as lipid-lowering agents for 20 years before this discovery, are weak PPAR α agonists, effective on human PPAR in the micromolar range, explaining the observation that they are given in the range of 100–1,200 mg per day. Fibrates, such as fenofibrate, mainly act via activation of PPAR α in the liver to regulate genes involved in fatty acid oxidation [19]. They were then called PPAR α activators, and their main laboratory effects are to reduce triglycerides and increase high-density lipoprotein (HDL) cholesterol levels. The first potent and selective PPAR α agonist acting in the nanomolar range with clinical data was LY518674, the development of which was stopped in 2007 when phase 2 studies showed no advantage over existing fenofibrate [20].
The link between PPAR γ activation and the thiazolidinedione insulin-sensitizing agents pioglitazone and rosiglitazone was established by researchers at Upjohn and Glaxo in 1994 and 1995, respectively [21, 22]. PPAR γ increases adipocyte differentiation and storage of fat. The short-term marker of PPAR γ activation in plasma is an increase in levels of the adipocytokine adiponectin, which increases insulin sensitivity in liver and muscle [23, 24]. The first animal results with PPAR δ agonists L165041 and GW501516 were reported in 1999 by researchers at Merck and in 2001 at Glaxo [25, 26].
The PPAR Machinery
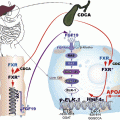
The PPAR machinery is similar to other nuclear receptors with sequential complexes of coactivators and corepressors with enzymatic activities (for review, see Rosenfeld 2006) [27] and a series of metabolic transformations that turn PPARs toward activation or direct them to degradation (Fig. 19.2). The role of these different proteins, their metabolic transformations, and the concept of selective PPAR modulator are summarized in the next sections. Without ligand the transcription of DNA into messenger RNA is usually repressed by the binding of corepressors on the heterodimer PPAR–RXR, and chromatin is compacted (Fig. 19.3). With the presence of ligand in the ligand binding domain, the structural changes in the AF-2 region permit to replace corepressors by coactivators, to associate remodeling of chromatin by acetylation of histones, in order for RNA polymerase to access the DNA and initiate transcription (Fig. 19.4). One important aspect common to PPAR activation is transrepression of inflammatory genes under the control of nuclear factor kappa B (NFκB) or activated protein (AP) 1. This transrepression is an indirect effect since there is no PPRE in the promoter. This was shown for PPAR γ on induction of tumor necrosis factor (TNF) α by phorbol myristate acetate in human monocytes/macrophages [28], for PPAR α on human aortic smooth muscle cells and interleukin (IL)-1-induced IL6 expression [29, 30], and for PPAR δ with expression of monocyte chemoattractant protein (MCP)-1 [31]. In human endothelial cells, fenofibrate and L165041, but not rosiglitazone, inhibited TNF α-induced monocyte adhesion, vascular cell adhesion molecule-1 (VCAM-1) expression, and monocyte chemotactic protein-1 (MCP-1) secretion through inhibition of nuclear P65 translocation, necessary for NFκB activation [32].
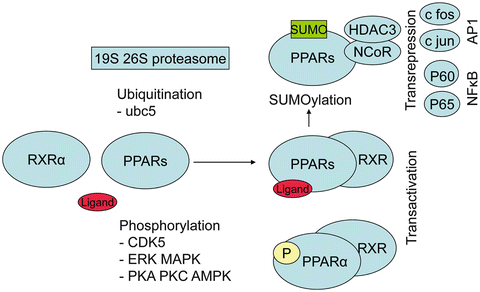
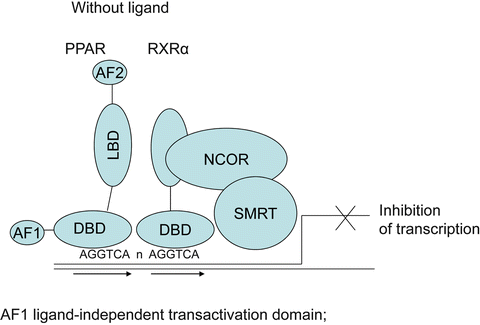
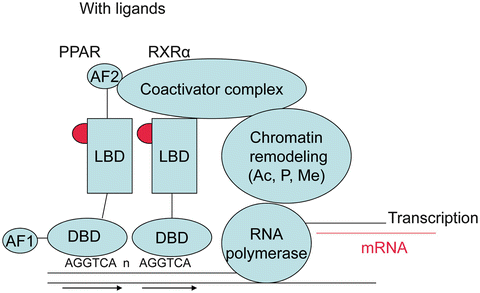

Fig. 19.2
PPAR network. Upon activation with ligand, PPAR heterodimerizes with RXR α and activate target genes (transactivation). Phosphorylation has opposite effect transactivation for PPAR α or its inhibition for PPAR γ. Sumoylation of PPAR is associated with transrepression which prevents transcription of NFκB or AP-1-dependent inflammatory genes and with a reduction of degradation in the proteasome. CDK5, cyclin-dependent kinase 5; ERK MAPK, mitogen-activated kinase; PKA PKC AMPK, protein kinase A or C and AMP-activated kinase; NCoR, nuclear corepressor; HDAC3, histone deacetylase 3

Fig. 19.3
Corepressor complex: without ligand, PPAR and RXR α are linked to their PPRE direct repeat (AGGTCA n AGGTT) by the DNA binding domain; the corepressors NCoR and SMRT prevent DNA transcription. AF1 AF2 ligand-independent transactivation domains 1 and 2; DBD, DNA binding domain; LBD, ligand binding domain; NCoR, nuclear corepressor; SMRT, silencing mediator for retinoid and thyroid hormone

Fig. 19.4
Coactivator complex: with fixation of ligands, conformational changes in ligand binding domain permit replacement of corepressors by coactivators, of which the enzymatic activities acetylate, phosphorylate, or methylate the chromatin allowing access to DNA or RNA polymerase and initiation of transcription into copies of messenger RNA
PPAR Coactivators and Corepressors
The main PPAR coactivator, or at least the best studied one, is peroxisome proliferator-activated receptor γ coactivator 1 α (PGC-1 α) [33]. Through a number of transcription factors, including PPARs, PGC-1 α modulates numerous metabolic pathways in the liver, skeletal and cardiac muscle, and adipose tissue, including gluconeogenesis and glycolysis, fatty acid synthesis, and oxidation. Indeed, PGC-1 α itself is subject to the same modulations as PPAR (see below through phosphorylation, ubiquitination, or sumoylation). Other PPAR coactivators are steroid receptor coactivator 1 (SRC1) and cyclic adenosine 5′-monophosphate (cAMP) response element binding protein (CBP/P300) which possess histone acetyltransferase activity, leading to the decondensation of chromatin which is necessary for gene transcription.
The main PPAR corepressors, nuclear receptor corepressor (NCoR) and silencing mediator for retinoid and thyroid hormone (SMRT), are associated with histone deacetylase activity which maintain chromatin in a compact state. The role of NCoR was studied by specifically knocking out its gene in mouse adipocytes (AKO) or muscle (MKO). MKO mice were able to run longer than normal mice [34]. AKO mice had higher insulin sensitivity in liver, muscle, and adipose tissue than normal mice, with limited additional effect of rosiglitazone since PPAR γ target genes were already derepressed by NCoR deletion [35]. The effects of rosiglitazone to cause hemodilution were the same in AKO and normal mice. In MKO mice, exercise capacity and mitochondrial oxidation are enhanced by the loss of a transcriptional cofactor in muscle cells through modulation of transcription factors that includes PPAR δ. SMRT is a protein structurally similar to NCoR, which possesses different receptor interaction domains (RID) for different nuclear receptors, called RID2 for PPAR or RXR or RID1 for retinoid acid receptor [36].
Phosphorylation
Phosphorylation of PPAR γ by extracellular signal-related kinase (ERK) 1 at serine 112 inhibits adipogenesis [37]. Phosphorylation of PPAR α on serine residues in the ligand-independent transactivation domain AF-1 in response to insulin increases transcription activity through dissociation of corepressors [38]. HMG-CoA reductase inhibitors (“statins”) have been shown to stimulate PPAR α transcription by reducing its phosphorylation in HepG2 cells, a synergistic effect with fenofibric acid [39]. Transcriptional activation of PPAR α by bezafibrate was dose-dependently increased by statins in human kidney 293T cells. In addition, concomitant administration of fenofibric acid and pitavastatin decreased the transactivation of NFκB induced by phorbol myristate acetate (PMA) [40]. Data on PPAR δ phosphorylation are limited to the location of predicted consensus phosphorylation sites and inhibition of PPAR δ activation by kinase inhibitors [41].
It was shown recently that phosphorylation of PPAR γ at serine 273 by activated CDK5 leads to a loss of transcription of PPAR γ in adipocytes [42]. The cyclin-dependent kinase (CDK) 5, which is present in the cytoplasm and the nucleus, is activated by phosphorylation at tyrosine 15 within a high glucose milieu and IL1β, by TNF α, or by high-fat diet. This finding permitted the same authors to discover new small molecules binding to PPAR γ blocking CDK5 serine 273 phosphorylation, like thiazolidinediones (TZDs), with potent antidiabetic activity in insulin-resistant mice fed with a high-fat, high-sugar diet, without causing fluid retention and weight gain [43].
Ubiquitination
Proteins are degraded in the proteasome after fixation on lysine residues of repeated sequences of a small 76AA polypeptide called ubiquitin. In the absence of their ligands, PPARs are rapidly degraded by this process. The degradation of PPAR γ is increased by different TZD ligands [44]; conversely, ubiquitination of PPAR α is reduced transiently with different fibrate ligands [45], and ubiquitination of PPAR δ is markedly reduced by PPAR δ agonists [46].
Sumoylation
Sumoylation is the attachment of another polypeptide of 101 amino acids called SUMO, for small ubiquitin-like modifier. Sumoylation at a lysine in the ligand binding domain of PPAR γ is the mechanism which converts activation of transcription by rosiglitazone into repression of NFκB or activator protein (AP) 1 in murine macrophages. This prevents ubiquitination of NCoR to maintain repression of inflammatory genes such as inducible NO synthase [47]. In adipose tissue, sumoylation of PPAR γ, which reduces the effect of rosiglitazone, is increased in the absence of the hepatokine fibroblast growth factor (FGF) 21 [48].
Similarly, sumoylation at lysine 185 has been identified in the hinge region of PPAR α [49]. To date, a potential sumoylation site for PPAR δ has been suggested on lysine 185, as for PPAR α.
Posttranslational regulation of PPARs by different patterns of mono- or polyubiquitination, as well as by mono- or polysumoylation, has been recently reviewed by Wadosky and Willis [50]. This review also reports that the coreceptor RXRα and the coactivators PGC-1α can be ubiquitinated or sumoylated, adding to the complexity of these regulatory processes.
Acetylation
Acetylation and deacetylation of genes are major processes affecting gene expression through decondensation and recondensation of chromatin. It also affects proteins. The first nuclear receptors shown to be acetylated were the androgen–estrogen receptors; this has not been shown clearly for PPAR [51]. However, their key coactivator PGC-1 α is inactivated by acetylation in high-energy states or deacetylated by sirtuin 1 in low-energy states [52]. The nicotinamide adenine dinucleotide (NAD)-dependent histone deacetylases or sirtuins by interacting with PPARs and their coactivators thus provide a new level of complexity to the regulation of nuclear receptors [53].
Partial Agonists or SPPARMs
A partial agonist is a ligand that induces a submaximal response even at full receptor occupancy. It can also reduce the full PPAR γ agonist response. For instance, in comparison with rosiglitazone, troglitazone is a full agonist on murine 3T3L1 adipocytes but a partial agonist in muscle C2C12 myotubes and HEK293T kidney cells [54]. Olefsky proposed to name selective PPAR modulators (SPPARMs); such products differ from full agonists by differential regulation of target genes [55]. SPPARMs are designed to separate efficacy and adverse effect dose–response curves. This concept was already developed in nuclear receptor pharmacology, with selective estrogen receptor modulators (SERMs), such as tamoxifen or raloxifene, which recruit corepressors such as NCoR to the AF-2 region, whereas estradiol recruits coactivators such as the glucocorticoid receptor-interacting protein 1 (GRIP1) [56] or with selective vitamin D modulators such as paricalcitol with differential recruitment of coactivators than calcitriol, the active form of vitamin D [57].
Increasing concentrations or doses with full PPAR γ agonists lead to greater efficacy but greater adverse events, such as weight gain and volume expansion.
PPAR γ partial agonists such as balaglitazone or INT131 displace a full agonist such as rosiglitazone. Metaglidasen, the (−) stereoisomer of halofenate, tested in the 1990s as a lipid-lowering agent, is another selective partial PPAR γ modulator and is still in clinical development for its uricosuric activity. They bind the same pocket as TZDs, which is required to block PPAR γ phosphorylation, but induce different conformational changes in PPAR γ, leading to different recruitment of coactivator/corepressor. As an example, INT131 induces less recruitment of DRIP205 (vitamin D-interacting protein 205), a coactivator involved in lipid accumulation than rosiglitazone or pioglitazone in HEK cells [58]. The same finding was reported with fibrates: gemfibrozil induced less recruitment of DRIP205 than fenofibrate and behaves as a partial agonist to increase apoA-I activation. This translated in a comparative trial in dyslipidemic patients to a larger increase in apoA-I levels, a protective apoprotein in HDL, with fenofibrate than with gemfibrozil [59].
Effects of PPAR Agonists in Diabetes
This review is limited to PPAR activators or agonists which entered clinical development in diabetes and/or dyslipidemia (Table 19.1). Few PPAR antagonists were synthesized and they were not developed for the treatment of diabetes [60]. GW6471, a potent PPAR α antagonist, is used as a pharmacological agent to test whether an effect is PPAR dependent or PPAR independent. GW9662 is a PPAR γ antagonist which promotes the recruitment of NCoR. Finally, GSK0660 and GSK3787 are PPAR δ antagonists for pharmacological use.
Table 19.1
Phase of clinical development reached by PPAR agonists
PPAR α | PPAR γ | PPAR α/γ | PPAR δ | Pan-PPAR | |
|---|---|---|---|---|---|
Marketed | Bezafibrate Ciprofibrate Fenofibrate Gemfibrozil Clinofibrate Etofibrate | Pioglitazone Rosiglitazone | |||
No more marketed | Clofibrate | Troglitazone | |||
Phase 3 | Balaglitazone Metaglidasena Rivoglitazonea Ciglitazone Farglitazarb | Aleglitazardd Lobeglitazone Muraglitazar Ragaglitazar Tesaglitazar Imiglitazar MK767 | |||
Phase 2 | K877 ZYH7 AVE8134 GW590735 KRP-105 LY518674 CP778875 | INT131 MBX2044 FK614 | Cevoglitazar Naveglitazar Sipoglitazar | MBX8025 GW501516 GW0742 L165041 | GFT505c Chiglitazar Indeglitazar Sodelglitazar Netoglitazone |
The organs implicated in glucose control are listed in Table 19.2. With their direct effects on gene expression and their indirect effects on inflammation, and according to their tissue distribution, PPARs affect most of these organs, beyond the liver for PPAR α, the adipose tissue for PPAR γ, and the skeletal muscle for PPAR δ. In the kidney, they have different locations: PPAR α is located mainly in the proximal tubule, the medullary thick ascending limb, and in the mesangium; PPAR γ in the distal medullary collecting duct and glomeruli; and PPAR δ in a diffuse fashion as in other organs [62]. In the brain, the interplay of PPAR subtypes has been shown in cultures of astrocytes, where the three subtypes are present. Combined application of PPAR γ and PPAR δ activators increased cyclooxygenase 2 expression induced by lipopolysaccharide, whereas the additional application of a PPAR α agonist abolished this effect [63].
Table 19.2
Organs implicated in glucose control
PPAR α | PPAR γ | PPAR δ | |
|---|---|---|---|
Liver | Increase in fat oxidation and apoA-1 increase insulin sensitivity | Decrease in steatosis Increase insulin sensitivity | |
Skeletal muscle | Increase insulin sensitivity | Increase in fat oxidation and energy expenditure | |
Adipose tissue | Reduction in inflammatory adipocytokines | Increase in adipocyte differentiation and adiponectin release | |
Pancreas | Amplification of glucose-induced insulin secretion | ||
Gut | Anti-inflammatory | Increase in GLP1 production | |
Vascular wall | Increase in NO synthesis |
In the pancreas, the three PPARs are expressed in pancreatic β cells. PPAR α modulates fatty acid oxidation and PPAR γ directs them toward esterification. Although PPAR δ is the most abundant PPAR in the pancreas at the mRNA and the protein level, until recently its effects on fatty acid oxidation have been less well studied [64]. PPAR δ activation increases fatty acid oxidation and to a larger extent than PPAR α activation. In the pancreas, fatty acids acutely potentiate glucose-stimulated insulin secretion (GSIS), but their chronic exposure elevates basal insulin secretion and alters GSIS, a phenomenon called lipotoxicity.
Discordant results are reported in the literature with PPAR α or PPAR γ agonists. PPAR α was described to potentiate and PPAR γ to attenuate GSIS in INS-1E cells, an immortalized insulinoma rat cell line [65]. On the contrary, in vivo, the PPAR α agonist fenofibrate impaired GSIS in neonatal rats receiving monosodium glutamate to induce obesity, while pioglitazone, a PPAR γ agonist, increased it in db/db mice [66, 67]. This discordance might be explained by the low expression level of PPAR γ in INS-1E cells.
Activation of PPAR δ by unsaturated FAs or a synthetic ligand enhanced GSIS in primary rat islets or INS-1E cells without affecting basal insulin secretion [64]. In order to maintain β cell function, PPAR δ would play a role of lipid sensor to adjust the mitochondrial fatty acid oxidation. It was recently suggested that 4-hydroxynonenal (4-HNE) was one endogenous activating ligand of PPAR δ [68]. The level of reactive oxygen species (ROS), such as 4-HNE, is essential to β cell function, as low-level ROS production increases glucose-induced insulin secretion, whereas high levels of ROS can induce β cell apoptosis.
GSIS is also linked to influx of calcium ions to the cytosol induced by depolarization from the voltage-dependent Ca++ channel. In INS-1 cells, the sarco-endoplasmic reticulum Ca++ ATPase (SERCA2) pump maintains intracellular Ca++ homeostasis, in particular, a high Ca++ level in the endoplasmic reticulum. The expression of this pump is decreased in animal models of diabetes and in diabetic human islets. Pioglitazone directly increases expression of SERCA2 through transcription of the gene and indirectly through prevention of CDK5-induced phosphorylation of PPAR γ [69]. This experiment suggests that blocking CDK5 could permit to dissociate positive effects on glucose homeostasis from other effects from PPAR γ agonists.
Effects of PPAR Agonists in Type 1 Diabetes
Clinical studies with PPAR agonists in type 1 diabetes (T1DM) are limited to their effects on lipid or glucose markers. One placebo-controlled randomized study was conducted with fenofibrate in 44 patients with T1DM to assess its effect alone or in combination with vitamin E for 8 weeks on copper-induced oxidation of LDL and VLDL particles [70]. The lag time of oxidation was significantly prolonged by fenofibrate 200 mg + vitamin E 400 IU. A placebo-controlled randomized study is in the planning stage to evaluate the effects of fenofibrate on progression of diabetic retinopathy in 400 adults with T1DM (http://clinicaltrials.gov/ct2/show/NCT01320345) [71]. The lipid-modifying effects of bezafibrate in T1DM were evaluated in earlier placebo-controlled studies [72, 73]. Of note, this fibrate, now considered as an archetype pan-PPAR agonist in transactivation assays, did not improve HbA1c after 3 months of treatment [40, 74].
Three placebo-controlled randomized studies have been reported with TZDs in T1DM patients on insulin therapy, with modest insulin-sparing effects as compared to those observed in T2DM. In 50 overweight adults with T1DM, an 8-month intervention to achieve glycated hemoglobin level of 7.0 % required an 11 % increase in the daily dose of insulin in the placebo group, but no change in the rosiglitazone group [75]. In 36 T1DM adolescents aged 10–18 years, the dose of insulin was increased by 9 % with placebo and reduced by 6 % with rosiglitazone after 6 months of treatment, with HbA1c remaining stable around 8.5 % [76]. In 60 lean T1DM patients aged 14 years or more, 6-month treatment with pioglitazone was associated with a significant decrease in HbA1c (0.2 %) and in postprandial glucose levels (0.7 mmol/L) in the intervention group only, with no changes in insulin doses [77]. In patients with slowly progressive T1DM, diagnosed by the presence of glutamic acid decarboxylase (GAD) antibodies, an insulin-requiring state defined by HbA1c and post-glucose C-peptide levels was reached at 4 years in 4/4 subjects randomized to pioglitazone as compared to 1/5 subjects randomized to metformin [78]. Thus, the effects of TZD in T1DM sharply differ from those reported for T2DM prevention with troglitazone in TRIPOD [79], rosiglitazone in DREAM [80], and, more recently, pioglitazone in ACT-NOW [81] and from their effects on glucose control in people with established T2DM.
Effects of PPAR Agonists in Type 2 Diabetes and Dyslipidemia
For the treatment of T2DM, the first TZD PPAR γ agonist troglitazone was introduced in the USA in October 1997 and was withdrawn in March 2000 for hepatic toxicity. Rosiglitazone and pioglitazone were introduced in the USA in 1999 and in Europe in 2000. In Japan, pioglitazone was introduced in 1999 and rosiglitazone in 2003. The effects of pioglitazone on macrovascular events in 5,238 T2DM patients were reported in 2005 [82]. Although the study primary end point was not reached, there was a significant 16 % reduction in the main secondary end point, which included death from any cause, acute nonfatal myocardial infarction, or stroke. The effect of TZDs on diabetes control and the controversy about their hazard on cardiovascular events have been the subject of numerous reviews in the last 5 years [83–85].
The first request for approval of a PPAR α/γ dual agonist, muraglitazar, was submitted to the Food and Drug Administration (FDA) for registration but was withdrawn in May 2006 after a combined analysis of clinical studies indicated an increased cardiovascular risk [86]. Such an increase in cardiovascular risk led to the suspension of registration of rosiglitazone in Europe in September 2010 and severe limitations to its use in the USA. Finally, in June 2011, pioglitazone was withdrawn from some European markets due to increased risk of bladder tumors, a decision not endorsed by the European Medicines Agency.
Discontinuation of the development of PPAR agonists occurred for multiple reasons: toxicity of the compound (vascular or bladder tumors in rodents with MK767 or ragaglitazar, respectively), long duration of development, clinical adverse events, expectation not to be better than existing drugs, and stopping of development efforts in the cardiometabolic domain. In particular, the FDA requested in July 2004 that 2-year rodent carcinogenicity studies be completed and reviewed before proceeding to phase 3 studies of more than 6-month duration. This decision was made after the evaluation of carcinogenicity in rodents for 11 PPAR agonists, with the observation of hemangioma/hemangiocarcinoma with 8/11 compounds and urinary bladder/renal pelvic transitional cell carcinomas with 5/6 PPAR α/γ dual agonists and pioglitazone (www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM071624.pdf) [87]. In addition, the FDA requested in December 2008 that new antidiabetic agents had to demonstrate through randomized, prospective clinical trials that they do not increase risk for cardiovascular events (www.fda.gov/downloads/Drugs/Guidance Compliance RegulatoryInformation/Guidances/UCM071627.pdf) [88]. The thiazolidinedione intervention with vitamin D evaluation (TIDE) study, a large intervention study to assess the effect of the existing TZDs pioglitazone and rosiglitazone on cardiovascular events, planned in 16,000 T2DM patients at risk of CVD events was initiated in 2009 but stopped by the FDA 1 year later leaving uncertainty about the risks and benefits from TZDs (TIDE 2012) [89]. The authors stated that, had this study been initiated earlier, it would have provided clear evidence regarding the efficacy and safety of rosiglitazone and pioglitazone.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree