Personalized medicine in oncology drug development
Nicholas C. Dracopoli, PhD  Iqbal Grewal, PhD, DSc, FRCPath
Iqbal Grewal, PhD, DSc, FRCPath  Chris H. Takimoto, MD, PhD, FACP
Chris H. Takimoto, MD, PhD, FACP  Peter F. Lebowitz, MD, PhD
Peter F. Lebowitz, MD, PhD
Overview
The concept of personalized medicine in oncology drug development is no longer an aspiration; it is now, in large measure, the accepted paradigm of how oncology drugs should be developed. In modern oncology drug development, determining which molecularly characterized tumors will benefit the most from a drug is a critical part of the development plan. In addition, another important personalized medicine approach is emerging whereby a therapeutic is specific to an individual patient. Examples of this individualized therapeutic approach include engineering of autologous immune cells with reinfusion into the patient or cancer vaccines that are specific to an individual tumor.
A growing number of successful examples have demonstrated that the personalized medicine approach via biomarker selection can provide more benefit to patients and more rapid approval of drugs. The benefits of a personalized medicine strategy in oncology drug development are multiple and include an increased probability of overall success, smaller Phase 3 registration trials, and the potential for enhanced clinical benefits for patients. In a recent review, Falconi and colleagues examined the outcome of 676 Phase 1, 2, and 3 clinical trials of novel therapeutics for non-small cell lung cancer conducted from 1998 to 2012.1 Overall, the cumulative success rate, as defined by the advancement to the next stage of clinical testing or to approval, was only 11%. However, when a biomarker was utilized for patient selection, the cumulative success rate increased by nearly sixfold to 62%. Thus, there is strong objective evidence for predictive biomarkers to enhance the probability of success in drug development. Using specific genetic and/or protein markers, a number of important drugs have been developed and approved in specific populations. This approach represents a paradigm shift that has proved so powerful that it is now a standard.
Introduction
The growing armamentarium of biomarker tools will allow a further expansion of the personalized drug development approach. It will also increase complexity across development and this will come with challenges. While precision medicine is now a reality, the successes have generally been where the companion diagnostic is the same as the target of the drug. As the science advances, efforts will need to be expanded in a few areas of personalized medicine to realize further gains. Some examples include
- 1. Multiplex biomarker panels as diagnostics. With few exceptions, most successful personalized medicine programs in oncology have measured a single gene or protein. As multiplexed approaches become mainstream with the analysis of thousands of datapoints, diagnostics using full panels will begin to emerge. This may require different clinical development strategies with the identification of a biomarker panel in early clinical trials and confirmation in registration studies.
Some successful examples of multiplex diagnostics include transcriptional profiling efforts. Using this approach, a gene expression pattern is identified for responders versus nonresponders. This profile is then prospectively applied to a validation set of samples to determine the predictive value, sensitivity, and specificity of the test. Of the profiles in clinical use, such as Oncotype Dx (Genomic Health, Redwood City, California), MammaPrint (Agendia BV, Amsterdam, the Netherlands), and H/I (AvariaDX, Carlsbad, California), these tests are better at identifying the patients unlikely to benefit from treatment than the potential responders.
- 2. Immuno-oncology personalized medicine. While analysis of cancer cells has formed the mainstay of personalized medicine, it is becoming increasingly clear that other components of a tumor, such as immune cells and stromal cells, are critical biologic mediators that can now be manipulated for therapeutic benefit. As new therapies target these microenvironment cells, new technologies for profiling are being developed. The complexity of measuring multiple markers in multiple cell types presents challenges in the development of novel immunotherapy.
- 3. Personalized therapeutics. These include engineered autologous cells and individual tumor antigen approaches and are discussed in detail below.
Thus, while personalized medicine has finally become a reality in oncology drug development, it is becoming a far more complex proposition as we address more complex mechanisms and biology. In this chapter, we try to cover some of the key issues that are critical to understand in order to achieve further successes.
Role of biomarkers and companion diagnostics in personalized medicine
Types of biomarkers
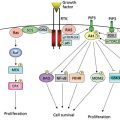
Biomarkers and their associated diagnostics are critical components of most personalized drug development efforts. Biomarkers can be classified into several types as shown in Table 1. These include predictive, prognostic, and surrogate end points. All of these biomarker types contribute in critical ways to drug development, but we typically think of predictive biomarkers as being the driving force in personalized medicine. These are used to identify patients within a population who have a higher probability of responding to treatment than the “all-comer” population. The symbiotic development of molecular profiling technologies and the completion of the human genome sequence created new opportunities for the comprehensive discovery of markers to help predict drug efficacy or reduce risk of drug toxicity.
Table 1 Types of Biomarkers and Diagnostics
| Marker | Function | Test |
| PD/MOA |
|
|
| Predictive |
|
|
| Resistance |
|
|
| Prognostic |
|
|
| Surrogate |
|
|
Today, an enormous number of biomarkers have been described, but most lack sufficient characterization necessary for clinical application. In October 2014, the GOBIOM database (www.gvkbio.com) lists more than 45,000 biomarkers. In effect, this is the equivalent of two biomarkers for every gene in the human genome! This suggests that the term biomarker is being greatly overused as we simply do not have this many useful biomarkers to support all different research applications. The vast majority of these biomarkers are poorly characterized and cannot be used effectively in research or clinical applications without considerable efforts to develop analytically valid assays as well as correlative data to confirm their role as a biomarker of a biological process.
Biomarkers have diverse uses throughout the drug development process as readouts for mechanism of action (MOA), pharmacodynamics (PDs), prognosis, response prediction, drug resistance, and surrogate markers (Table 1). Perhaps the most common use is as MOA markers to confirm the drug is hitting the expected target and having the desired downstream effect, often by measuring protein phosphorylation in members of the downstream signaling pathway. PD biomarkers are widely employed with pharmacokinetic (PK) analyses to determine a biologically effective dose and define the dose for the first Phase 2 efficacy studies. Prognostic markers are developed to determine the likely course of disease in the absence of any specific pharmaceutical intervention and have been widely used in some oncology indications to predict the likelihood of disease recurrence after resection of the primary tumor (e.g., Mammaprint and Oncotype Dx). Predictive biomarker tests are used to predict response to a specific treatment and are discussed in detail below. Increasingly, biomarker strategies are being used to detect and follow emergent resistance to therapy and considerable success has been achieved for the class of tyrosine kinase inhibitors to predict resistance to therapy.
Successes and challenges with companion diagnostics
As a testament to the successes of personalized drug development in oncology, as of January 2015, a total of 20 companion diagnostic tests have been approved by the FDA and are broadly used today in the treatment of cancer patients.2 These 20 tests are described in Table 2. Several important factors emerge upon close examination of the approved companion diagnostics in this table. Firstly, all the tests are simple and measure the status of the drug target or, for Kirsten-Ras (KRAS), a resistance pathway downstream of the drug target.3, 4 Each test is a single analyte test that measures the somatic genetic changes that activate the drug target including gene amplification (HER2, KIT), mutation (KRAS, epidermal growth factor receptor (EGFR), BRAF) or translocation (ALK). Secondly, all of the approved companion diagnostics in oncology are for signal transduction inhibitors with driver mutations in the target or downstream pathways. In contrast, there are no FDA-approved companion diagnostics for other drug mechanisms including cytotoxic drugs, epigenetic modulators, or immuno-oncology therapies. Why is it that we have been successful in developing companion diagnostics for signal transduction inhibitors but not for other important oncology drug targets? The answer is mainly that these driver mutations are the target (or closely related to the target) of the drugs being developed. Consequently, it is possible to quickly test the hypothesis that proliferation and metastasis of a tumor are driven by somatic abnormality detected in the drug target and that only those patients with such a somatic aberration will respond to inhibition of this target. This hypothesis can begin to be confirmed or refuted in clinical studies as soon as an efficacious dose is established in Phase 1 studies. In contrast, if samples have to be collected in Phase 2 for molecular profiling in order to develop a hypothesis, the development of a predictive marker becomes much more difficult. In this scenario, a stepwise approach must be taken which includes biomarker hypothesis generation, hypothesis testing and validation, and, finally, diagnostic development.
Table 2 List of approved FDA companion diagnostics in October 2014
| TEST | Manufacturer | Drug(s) | Method | Analyte |
| therascreen KRAS RGQ PCR Kit | Qiagen | Erbitux (cetuximab) | PCR | KRAS |
| — | — | Vectibix (panitumumab) | — | — |
| EGFR PharmDx Kit | DAKO | Erbitux (cetuximab) | IHC | EGFR |
| — | — | Vectibix (panitumumab) | — | — |
| therascreen EGFR RGQ PCR Kit | Qiagen | Gilotrif (afatanib) | PCR | EGFR |
| C-KIT PharmDx | DAKO | Gleevec (imatinib mesylate) | IHC | KIT |
| INFORM HER-2/NEU | Ventana Medical Systems | Herceptin (trastuzumab) | FISH | HER2 |
| PATHVISION HER-2 DNA Probe kit | Abbott Molecular | Herceptin (trastuzumab) | FISH | HER2 |
| PATHWAY ANTI-HER-2/NEU (4B5) | Ventana Medical Systems | Herceptin (trastuzumab) | IHC | HER2 |
| INSITE HER-2/NEU Kit | Biogenex Laboratories | Herceptin (trastuzumab) | IHC | HER2 |
| SPOT-Light HER2 CISH Kit | Life Technologies | Herceptin (trastuzumab) | CISH | HER2 |
| Bond Oracle HER2 IHC System | Leica Biosystems | Herceptin (trastuzumab) | IHC | HER2 |
| HER2 CISH PharmDx Kit | Dako Denmark A/S | Herceptin (trastuzumab) | CISH | HER2 |
| INFORM HER2 DUAL ISH DNA Probe Cocktail | Ventana Medical Systems | Herceptin (trastuzumab) | CISH | HER2 |
| HERCEPTEST | Dako Denmark A/S | Herceptin (trastuzumab) | IHC | HER2 |
| — | — | Perjeta (pertuzumab) | — | — |
| — | — | Kadcyla (ado-trastuzumab emtansine) | — | — |
| HER2 FISH PharmDx Kit | Dako Denmark A/S | Herceptin (trastuzumab) | FISH | HER2 |
| — | — | Perjeta (pertuzumab) | — | — |
| — | — | Kadcyla (ado-trastuzumab emtansine) | — | — |
| THxID BRAF Kit | bioMerieux | Mekinist (trametinib) | PCR | BRAF |
| — | — | Tafinlar (dabrafenib) | — | — |
| cobas EGFR mutation test | Roche Molecular Systems | Tarceva (erlotinib) | PCR | EGFR |
| VYSIS ALK Break Apart FISH Probe Kit | Abbott Molecular | Xalkori (crizotinib) | FISH | ALK |
| COBAS 4800 BRAF V600 mutation test | Roche Molecular Systems | Zelboraf (vemurafenib) | PCR | BRAF |
| BRACAnalysis CDx™ | Myriad Genetic Laboratories, Inc. | Lynparza™ (olaparib) | PCR | BRCA1/2 |
| Ferriscan | Resonance Health Analysis Services Pty Ltd | Exjade (deferasirox) | MRI | Liver iron |
Source: Data from http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm301431.htm.
One recent example of this complexity is seen with the development of immuno-oncology agents. Checkpoint inhibitors like anti-PD-1 antibodies have demonstrated significant survival benefits and are the vanguard of a new wave of immune modulators.5 These new drugs function by inhibiting immune checkpoints and preventing the tumor from evading the host immune system. However, early clinical data show that measuring the expression of the target checkpoints is quite complex as a predictive biomarker. While the response rate can be improved by—three to four times with high expression of PDL1 when using a PD1 inhibitor, it is also true that tumors without detectable expression respond to the drug. Consequently, simply testing for the expression of the checkpoint target may be insufficient as a predictive marker for this new type of drug and we may have to develop new molecular profiles to predict response to these important immune-modulating drugs.
In addition to the FDA-approved companion diagnostics for oncology drugs, there are many more tests being used for research and clinical applications in Clinical Laboratory Improvement Amendment (CLIA) and College of American Pathology (CAP) accredited laboratories. These tests vary from single-analyte tests to complex molecular profiles. Some have highly validated analytical performance and strong evidence of clinical utility, while other tests are much less well characterized. Recently, many laboratories have developed sequencing panels to screen for canonical cancer mutations in 30–300 genes. These panels are mostly used for research purposes and increasingly for screening cancer patients for enrollment into targeted clinical studies. Perhaps the best known of these tests is FoundationOne, provided by Foundation Medicine Inc. This test sequences 230 cancer genes in formalin-fixed paraffin-embedded tissue (FFPET) to identify somatic mutations. This information is analyzed and may be used by physicians to direct patients to appropriate clinical trials with targeted agents or, sometimes, off-label treatment with approved agents.
Strategies for companion diagnostic development
Companion diagnostic (CDx) development—preclinical approach
The first step in the CDx development is the formulation of the predictive biomarker hypothesis. Ideally, this begins at the same time a new therapeutic agent or target is identified. The predictive biomarker hypothesis outlines a strategy for identifying and testing assays that can prospectively predict drug sensitivity or resistance. Beginning at the earliest stages of drug discovery, this hypothesis should incorporate a thorough understanding of target biology and the drug’s proposed MOA. The resulting output is a series of candidate predictive biomarker assays that will undergo rigorous preclinical and clinical evaluation testing their value as CDx tests. Candidate biomarkers may predict either drug sensitivity or resistance. In either case, the CDx co-development strategy is similar.
As discussed above, in some programs, the predictive biomarker hypothesis is inherently defined by the drug target. For example, a program targeting a tumor resistance mutation might use the presence of that mutation as a predictive biomarker to select patients for early clinical trial evaluation to demonstrate rapid proof of concept. This approach has been utilized in the development of the second generation of EGFR inhibitors that target the T790M resistance mutation in NSCLC patients.6 In other situations, a predictive biomarker might simply be high expression of the drug target such as HER2/Neu expression in breast cancer patients optimally treated with trastuzumab.7 Most of the currently approved oncology CDx assays measure a single gene or protein directly related to the drug’s molecular target.
The specific characteristics of the proposed predictive biomarker test can influence its clinical validation. For example, a biomarker with a binary readout, such as the presence or absence of a mutation, is the simplest to evaluate in clinical trials because of the lack of ambiguity about what constitutes a positive test. In contrast, a continuous variable, such as intensity of immunohistochemical staining or the degree of gene amplification or copy number, is more complex because of the need to establish a threshold for test positivity in actual clinical studies. Even more complicated are multivariate tests, such as microarray signatures or composite parameters derived from a variety of different tests. In general, as these predictive biomarkers grow more sophisticated, the burden to conduct more highly powered clinical trials for clinical validation also increases.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree