Factors That Affect Therapy for Treatment of COVID-19
Symptoms
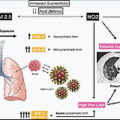
Symptoms of COVID-19 are many , ; some of them are as mild as those of normal influenza and others quite severe. Treatment provided to a patient with mild COVID-19 symptoms may be different from given to one with more serious symptoms such as difficulty of breathing. COVID-19 may lead to a few other manifestations such as cardiac problems in critically ill patients.
Most patients experience mild to moderate respiratory illness among other symptoms and recover quickly without requiring any special treatment. Aged patients and those with prior medical issues such as cardiovascular disease, diabetes, chronic respiratory disease, and cancer are at risk for developing serious complications and other manifestations. Several chapters in this book deal with varied manifestations, indicating that COVID-19 treatment is about not only respiratory illness but also potentially other severe symptoms, including tissue damage.
Disease Progression and Severity
In the time course of COVID-19, there are three stages: (1) early infection phase, (2) pulmonary phase, and (3) hyperinflammation phase. , It is important to note that some patients have only mild symptoms associated with an upper respiratory tract infection, or stage 1, whereas others progress to more advanced stages. The medication varies across stages. Pneumonia sets in at the pulmonary phase. Stage 3, or the hyperinflammation phase, leads to organ failures. Avoiding a cytokine storm remains a critical challenge in the progression of the disease.
Prevention and Control of Infection
To slow transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) one needs to know about the virus, the disease itself, and how it could spread. Social distancing and washing hands or using an alcohol-based rub frequently go a long way to avoid spread of the virus. SARS-CoV-2 spreads primarily through droplets of saliva or discharge from the nose when an infected person coughs or sneezes, which is why wearing face masks and social distancing have been shown to be effective against the spread of the virus. The transmission of SARS-CoV-2 also can be airborne.
In addition to the previously mentioned basic protective measures, when infection is suspected, identification of certain biomarkers becomes necessary for COVID-19 treatment. To form a therapeutic strategy, it is important to classify severity based on identified immune-related biomarkers so that appropriate treatment would be provided. A list of such biomarkers includes the following types:
- 1.
Hematological (lymphocyte count, neutrophil count, neutrophil-to-lymphocyte ratio [NLR])
- 2.
C-reactive protein (CRP)
- 3.
Erythrocyte sedimentation rate (ESR)
- 4.
Procalcitonin (PCT)
- 5.
Interleukin-6 (IL-6)
- 6.
Biochemical ( d -dimer, troponin, creatine kinase [CK], aspartate aminotransferase [AST]), especially those related to coagulation cascades in disseminated intravascular coagulation (DIC) and acute respiratory distress syndrome (ARDS)
Emerging Therapeutic Treatments of the Disease
Among the emerging therapeutic treatments for COVID-19, vaccines play an important part in preventing infection, but they are discussed elsewhere in a separate chapter in this book.
Among the many drug discovery efforts for COVID-19, drug repurposing has taken central stage thus far in the small molecule space wherein attempts have been made to repurpose drugs approved for other diseases into drugs for COVID-19. Some of these drugs have been accorded emergency use authorization and the repurpose drug remdesivir has been granted approval by the US Food and Drug Administration (FDA). A list of emerging treatments for COVID-19 includes use of various repurpose drugs, neutralizing monoclonal antibodies (mAbs), and cell and gene therapy covering convalescent plasma and stem cells. Most of these treatments are of tentative nature, some having greater promise than others. Further investigations continue in search of newer and more promising treatments for COVID-19. Countless journal articles, clinical practice guidelines, and other scientific reports on COVID-19 treatment have appeared in the last 18 months. Many clinical trials to evaluate promising repurpose drugs are also progressing. Given that mortaltiy of severe COVID-19 in unvaccinated persons remains high despite current treatment options, identification of newer targets and newer therapies for treatment of COVID-19 continues to be an important challenge in medical science.
Brief Description of Structure and Parts of SARS-CoV-2
Information on virus genome, structure, entry, replication, and transmission is covered in detail in Chapter 2 . SARS-CoV-2 is an RNA virus, and, once sequenced, the protein products of the viral genome can be determined. , The proteins are encoded through codons of three nucleotides. Each codon encodes for a particular amino acid, and the resulting amino acids are polymerized together to form a protein. The start and stop codons decide the beginning and termination of the polymerization process of the protein. From the genome, one can obtain the encoded protein products. The functions and structure of these proteins can be determined using bioinformatics approaches, which are discussed in a subsequent section.
The structural and functional details of a druggable proteome of SARS-CoV-2 have been determined through various bioinformatics and experimental approaches, and a detailed description is given elsewhere. Approximately two-thirds of the viral RNA genome of SARS-CoV-2 is translated into what are known as nonstructural proteins (NSPs). Besides this, the proteome encoded by the viral genome involves accessory proteins and four essential structural proteins that contribute to the stability and assembly of the virus, and they are the spike (S) receptor binding glycoprotein, which is the protein responsible for binding with the host angiotensin-converting enzyme-2 (ACE2) receptor and consequently plays a key role in the viral entry. For this discussion the nucleocapsid protein will be referred to with the symbol (N); the membrane protein will be referred to with the symbol (M), which is a transmembrane protein involved in the interaction with N, which will be referred to with the symbol (TM); and a small envelope protein that plays a vital role in assembly and stability of the virus will be referred to with the symbol (E). The functionality of SARS-CoV-2 can be broadly summarized as follows. The nonstructural proteins are predominantly involved in the viral replication process inside the human cell. The structural proteins referred to as M, N, S, and E proteins participate in viral assembly and stability. Other proteins known as accessory proteins interact with the host in functions such as shutting down host functions to redirect resources to viral replication, avoiding immune responses, and inducing pathogenicity. One of the compelling therapeutic strategies against COVID-19 is to inhibit the function of these proteins essential to the viral cycle of SARS-CoV-2 to stop both entry and replication of the virus, as described in the following sections.
Therapeutic Targets for the Treatment of COVID-19
The strategy for a therapeutic attack is twofold: virus-targeted antiviral therapy and host-directed antiviral therapy ( Fig. 17.1 ).

In virus targeted antiviral therapies, the targets are certain virus domains to block entry of virus into the human cell and the viral enzymes produced by the virus to hijack the host machinery for its survival and the replication of the virus.
In host-targeted antiviral therapy, the targets are receptors and proteins in the host whose function is critical for the virus to enter and hijack the host machinery for its replication and drug targets specific to different adverse symptoms caused by COVID-19. The adverse symptoms of COVID-19 are caused primarily by the unregulated immune response of the body.
Elements of both virus-targeted and host-targeted therapies are important for the treatment of COVID-19. The two types of therapies in the context of disease progression are shown in Table 17.1 . No immunosuppressants are used in stage 1 as the body’s natural resistance to the virus would be affected at that stage. No antivirals are used in stage 3. Immunomodulators or suppressants are required at both stages 2 and 3.
| Stage of Disease Progression | Virus-Targeted Therapy | Host-Targeted Therapy |
|---|---|---|
| Stage 1: Early viral infection | 1. To prevent viral entry into cell: Example: Specific monoclonal antibodies 2. To prevent viral replication: Antivirals Example: Remdesivir or Favipiravir | None |
| Stage 2: Pulmonary infection—susceptibility to develop pneumonia | Antiviral to lessen viral replication. | Immunomodulator ( example: dexamethasone) + anticoagulants |
| Stage 3: Hyperinflammation—acute respiratory syndrome (ARDS), sepsis, organ failures etc. | — | Immunomodulators + anticoagulants + other standard care |
Human genetic diversity introduces much more complexity to the COVID-19 disease progression and treatment than what is depicted in Table 17.1 . Many repurpose drugs are being used as part of either virus-targeted therapy or host-targeted therapy ( Tables 17.3 and 17.4 ).
| Target NSP | Target Function |
|---|---|
| NSP1 | Degrades host mRNA, inhibits IFN signaling |
| NSP2 | Degrades host mRNA, inhibits IFN signaling |
| NSP 3 (PLpro) | Degrades host mRNA, inhibits IFN signaling, cleaves polyprotein |
| NSP4 | Required for replication, DMV morphology formation with NSP3 |
| NSPp5 (Mpro) | Cleaves polyprotein |
| NSP6 | DMV morphology formation with NSP3 and NSP4 |
| NSP7 (RdRp) | Forms hexadecameric complex, processivity clamp RdRp and Primase complex |
| NSP8 (RdRp) | Forms hexadecameric complex, processivity clamp RdRp and primase complex |
| NSP9 | RNA binding |
| NSP10 | Stimulates activity of NSP14 and NSP16 |
| NSP12 (RdRp) | RdRp, virus replication and transcription |
| NSP13 | Virus mRNA capping, RNA helicase, 5′-triphosphatase |
| NSP14 | Virus mRNA capping and proofreading: 3′5′-exoribonuclease |
| NSP15 | Endoribonuclease, favors cleavage of RNA |
| NSP16 | Virus mRNA capping |
| Function | Repurpose Drug | Target/Action Related to COVID-19 |
|---|---|---|
| Antiviral | Favipiravir a : Active form is derived from nucleoside metabolite | RdRp polymerase inhibitor |
| Antiviral | Remdesivir: Prodrug of active form has nucleoside structure | RdRp polymerase inhibitor |
| Antiviral | Sofosbuvir/daclatasvir:Sofosbuvir—synthetic nucleoside drugDaclatasvir—nonnucleoside drug | Sofosbuvir: RdRp inhibitor. In HCV treatment, daclatasvir binds to NSP5A and prevents replication. In COVID-19, the mechanisms are expected to be similar b |
| Antiviral | Ribavirin: Synthetic nucleoside drug | Mechanism not established. Could be by RdRp polymerase inhibition |
| Antiviral | Molnupiravir: Synthetic nucleoside | RdRp polymerase inhibitor. c Drug under investigation |
| Antiviral | Oseltamivir | Neuraminidase inhibitor. Not considered effective against SARS-CoV-2 a |
| Antiviral | Darunavir | 3CLpro protease inhibitor. Not effective against SARS-CoV-2 a |
| Antiviral | Lopinavir/ritonavir | 3CLpro protease inhibitor. Not effective against SARS-CoV-2 a |
| Antiviral | Nelfinavir | 3CLpro protease inhibitor. Effective in vitro. More studies underway. Combination drug with virus entry inhibitor, cepharanthine d |
| Prevention of viral entry | Neutralizing monoclonal antibodies: (not repurposed) cocktail of (1) casirivimab and imdevimab and (2) sotrovimab | Binds to RBD of spike protein |
| Prevention of viral entry | Cepharanthine, an experimental alkaloid having macrocyclic structure | Predicted to bind to RBD domain of spike protein e |
a Parastoo T, Eftekhari S, Chizari M, et al. A review of potential suggested drugs for coronavirus disease (COVID-19) treatment. Eur J Pharmacol. 2021;895:173890. https://doi.org/10.1016/j.ejphar.2021.173890 .
b Sacramento CQ, Fintelman-Rodrigues N, Temerozo JR, et al. In vitro antiviral activity of the anti-HCV drugs daclatasvir and sofosbuvir against SARS-CoV-2, the aetiological agent of COVID-19. J Antimicrob Chemother. 2021;76(7):1874-1885. https://doi.org/10.1093/jac/dkab072 .
c Kabinger F, Stiller C, Schmitzova J, et al. Mechanism of molnupiravir-induced SARS CoV-2 mutagenesis. bioRxiv 2021.05.11.443555. doi: 10.1101/2021.05.11.443555 .
d Ohashi H, Watashi K, Saso W, et al. Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. iScience 2021;24(4):102367. https://doi.org/10.1016/j.isci.2021.102367 .
e Rogosnitzky M, Okediji P, Koman, I. Cepharanthine: a review of the antiviral potential of a Japanese-approved alopecia drug in COVID-19. Pharmacol Rep. 2020;72:1509-1516. https://doi.org/10.1007/s43440-020-00132-z .
The following is a brief discussion on what drug targets are and current approaches being followed to identify them. Most drug targets are proteins. Proteins are involved in many essential functions in living beings. Noted German researcher Paul Ehrlich once said, “drugs will not act unless they are bound” to protein targets. Drug targets are those proteins whose function or nonfunction and/or their overexpression or underexpression are causing the disease. Small or large therapeutic molecules can bind to binding cavities of these proteins and induce conformational changes within the proteins. Such changes could inhibit or activate the function of protein to restore normalcy of function from the disease condition of the body. The molecules that inhibit the function of proteins are called inhibitors or antagonists; the molecules that activate the function are called agonists. A sound understanding of the molecular-level mechanism of the disease leads to effective identification of the proteins involved in the molecular pathways and mechanisms of the disease and proteins with binding cavities that can qualify as drug targets.
Experimental Studies to Identify Druggable Targets
A target is called “druggable” if its activity (or function) can be modulated by a therapeutic drug. Proteins and nucleic acids are both examples of biological targets. A potential drug target should have druggability, have an acceptable toxicity profile, and be assayable in addition to the fact that its three-dimensional (3D) structure is known. There are two classes of methods for identification of targets, target deconvolution and target discovery. A gamut of experimental methods to identify targets are known both in vitro and in vivo and described in a seminal reference. The experimental methods include various affinity methods and genetic methods. Affinity chromatography is one of the most used methods for target deconvolution. Target deconvolution means looking for a target for an active compound. This results in screening of many molecules. In the target discovery method, the search is for a new target for which there is not yet a drug compound. Various biological assays and bioinformatics approaches are used for target discovery and target-based drug discovery.
Biological Assays
To understand the molecular mechanisms of disease, scientists perform experiments, specifically assays. In vitro assays are cell-based models, and in vivo assays are carried out inside living systems. Many different assays are used for drug target identification and validation; a few of them are mentioned here as examples. One major class of assays used in target identification and validation is functional assays, which are experiments designed to determine the role or function of a protein in a biological process or pathway.
As an example, to determine the activity of the target RNA-dependent RNA polymerase (RdRp) (NSP12), typical assays include the following 14 :
- 1.
Polymerase elongation template element (PETE)
- 2.
Fluorescence-based alkaline phosphatase–coupled polymerase assay (FAPA)
- 3.
Fluorometric RdRp activity assay
- 4.
Scintillation proximity assay
- 5.
Cell-based assays
When these assays are carried out in the presence of an antagonist, a decrease in the replication activity may be observed. To find the efficacy of the drug remdesivir, which has been approved by FDA for treatment of COVID-19, such assays were used to determine its inhibitory activity against RdRp.
Bioinformatics Approaches
Traditionally, in bioinformatics approaches, algorithms such as BLAST are used, to compute similarity of a given protein against proteins with a known structure and function present in bioinformatics databases. Through this process, one can predict the structure and function of a given protein by comparing its similarity with known targets. In every drug discovery project, one is also able to leverage and capitalize on the data available from experiments carried out previously, aided by bioinformatics, which is being further enhanced by advanced data-driven techniques. In the case of SARS-CoV-2, much of the knowledge from other virus systems such as SARS-CoV, Ebola, and Middle East respiratory syndrome coronavirus (MERS-CoV) led to recognition of similar target types for SARS-CoV-2.
Bioinformatics not only provides historical experimental data on structure, activity, and other properties of biomolecules but also gives insights from analysis of the data. Further, it provides many approaches not only for identification of targets but also for drug discovery. Current trends in bioinformatics encompass an approach that integrates many factors such as predicting disease-relevant genes, constructing gene networks, protein–protein interaction networks, biological pathway analysis, etc.
In silico or computational techniques, including molecular modeling techniques, can provide information on a variety of factors, such as:
- 1.
Binding orientation of small molecule inhibitors with protein structures in 3D space through molecular docking techniques
- 2.
Information on functional groups such as amino acid residues and other moieties that bind to the drug
- 3.
Stability and properties of ligand–protein complexes using molecular dynamics simulation
A combination of these techniques as part of an integrated approach guide the researcher in discovering new drug molecules and new drug targets. A summary of methods of target and drug discovery is given in Box 17.1 .
Step 1 : Identify differentially expressed proteins in the disease.
Step 2 : Use bioinformatics and AI a approaches to identify structure and function of those proteins.
Step 3 : Druggability analysis of the proteins.
Step 4 : Functional assays to experimentally validate the druggable protein target.
Step 5 : Select an experimentally validated target for drug discovery.
Step 6 : Learn binding pocket information from crystal structures of protein-ligand complex.
Step 7 : Use molecular modelling and AI approaches to screen hits (either repurpose drugs or NCE b s) by molecular docking of drug molecules in the binding pocket.
Step 8 : Prioritize top hits using calculated docking scores or binding energies.
Step 9 : In Vitro/In Vivo evaluation of safety and efficacy to identify lead molecules.
Step 10 : Clinical trials of the drug lead candidates to identify active drug molecule.
a AI stands for “artificial intelligence” approaches, where using trained mathematical models, structure and properties are predicted;
b NCE stands for “new chemical entity” which means new drug molecule.
The literature published in the last 18 months reveals the extensive use of various bioinformatics approaches related to COVID-19. , Bioinformatics enables scientists to reduce the number of experiments and quicken the target discovery, drug discovery, or drug repurposing process. Drug repurposing is particularly attractive because the knowledge of the drug safety is already available, which saves time and reduces cost.
In addition to the previous description, it is important to point out that bioinformatics approaches are enhanced by integration of various data. Genomics, transcriptomic, and metabolomic data are integrated with proteomics (proteins) data through data science techniques to make predictions about protein biomarkers of a disease. The predictive modeling method helps narrow the search space of druggable protein targets from a list of proteins involved in the disease pathway. The druggable proteins identified should be unique to the disease to avoid off-target effects.
To identify the differential expression of genes and their protein products in the diseased and the normal case, various forms of high-throughput expression profiling is carried out and data are deposited in many public research databases, such as the National Center for Biotechnology Information Expression Atlas. , Various forms of data analysis–based machine learning algorithms are used to identify the differentially expressed genes, which are then mapped to their respective protein products. The protein products that are identified with druggable pockets and unique differential expression with respect to the disease are proposed as possible therapeutic targets.
Further, functional assays are carried out to validate a proposed target. If target protein inhibition is proposed as a therapeutic strategy to overcome the overexpression of the target protein in the disease, gene knockout experiments are performed, in which the gene corresponding to the target is knocked out and thus the target protein is not expressed; if an attempt to induce the disease fails in such a case, the target can be considered and experimentally validated. Although experimental methods are still required to validate proposed targets, the machine learning and bioinformatics approaches are useful to carry out the data analysis required to identify perturbed molecular pathways in a disease, which leads to identifying novel therapeutic targets. A dedicated resource of COVID-19–related differential gene expression data is available at CovidExpress for the leveraging of bioinformatics and machine learning approaches in identifying novel therapeutic targets.
A recent example of identifying a drug target and repurposing a known drug molecule for COVID-19 is related to baricitinib, a known immunomodulator. Baricitinib is an oral Janus kinase (JAK)1/JAK2 inhibitor approved for the treatment of rheumatoid arthritis. Based on a large repository of structured medical information extracted from the scientific literature using machine learning, scientists identified JAK1/JAK2 and AP2-associated protein kinase 1 (AAK1) proteins as potential targets for treatment of COVID-19. , AAK1 is known to regulate endocytosis. Further, it was predicted through modeling studies that baricitinib would bind to the AAK1 protein as well, which would inhibit virus entry. A combination of baricitinib with an antiviral drug (remdesivir) is in clinical testing. Thus a multifaceted approach comprising experimental, theoretical, computational, and data science–based techniques is used to identify drug targets and drug molecules for COVID-19.
The host genetic diversity that determines susceptibility to SARS-CoV-2 and disease severity makes a case for the use of precision medicine methodology in COVID-19 therapeutics. One of the challenging problems in COVID-19 treatment, for example, is the knowledge gap of predicting which patients have a genetic predilection for a cytokine storm and thus would benefit most from immunosuppressive medication such as corticosteroids. To deal with this challenge, Lam C. et al. used a precision medicine–based artificial intelligence (AI) a
a AI stands for artificial intelligence approaches, in which by the use of trained mathematical models, structure and properties are predicted.
model to make recommendations on the use of corticosteroids for COVID-19.Virus-Targeted Antiviral Therapy: Druggable Nonstructural Proteins
In this section, the functions of the nonstructural proteins of SARS-CoV-2 are briefly described to indicate why some of these proteins are targets to treat COVID-19. Table 17.2 provides a summary of the various NSPs and their functions. The idea has been to inhibit functions of these NSPs by binding them with small molecules of appropriate structure. The function of NSP11 is yet not understood.