The Oral Cavity and Viral Infections
The oral cavity is home to a vast microbial flora that also include a robust community of viruses. Although the viruses that infect bacteria or bacteriophages constitute the majority, viruses that infect human cells form a significant component of the oral microbiome. , Much like bacteria, viral communities are also site-specific alterations that can lead to disease. Indeed, one of the earliest associations of biofilm virome with human disease is the increased membership of lytic bacteriophages in subgingival plaque in periodontal disease. Although the evidence for oral virome and the contributions to oral health and disease is still emerging, the proclivity for specific viruses to affect the oral cavity is well recognized. , Furthermore, direct communication between the oropharynx and the nasopharynx makes the highly vascularized oral tissues more vulnerable to viruses harboring the respiratory or gastrointestinal (GI) tract. Consequently, many of these viruses have been implicated in the cause of chronic oral mucosal lesions. , In the sections that follow, we briefly review the oral manifestations of major DNA and RNA viruses, discuss the SARS-CoV-2 infection with particular focus on the pathology of oral symptoms, debate the scope of salivary diagnostics for SARS-CoV-2 infections, and, finally, the potential of SARS-CoV-2 transmission through saliva, including the spread in dental offices.
Viral Infections With Significant Orofacial Manifestations
DNA Viruses
The human herpesvirus (HHV) family constitute the most common viral infections of the oral cavity. They are relatively large, enveloped, double-stranded DNA viruses with icosahedral symmetry. Of the eight members in the family, the herpes simplex virus-1 (HSV-1) infection is the most common and is characterized by painful mucosal ulcers affecting the attached gingiva, such as the hard palate and the gum tissues surrounding the teeth and the portions of the jawbones encasing them. Following primary infection, the HSV typically remains latent in sensory nerve ganglia, and a recurrence occurs when the virus is reactivated, often seen clinically as herpes labialis characterized by periodic appearance of cold sores or ulcers in the lip. Infections by varicella zoster virus (VZV) and Epstein Barr virus (EBV) viruses are the next common HHV infections. VZ causes chickenpox that could manifest with blue Koplik spots in the oral mucosa, and the EB virus causes infectious mononucleosis (the kissing disease). Human papillomavirus (HPV) is a small, nonenveloped, double-stranded DNA virus with icosahedral symmetry and more than 70 genotypes. HPV 16 and 18 are often associated with benign papillomatous lesions of the oral mucosa. In addition, focal epithelial hyperplasia, or Heck disease, and condyloma constitute oral HPV infections. , , A significant finding is that HHVs are established risk factors for oral cancers and precancerous lesions. This role has been variably attributed to the virus-induced dysbiosis, increased inflammation, and production of carcinogenic metabolites.
RNA Viruses
Among the retroviruses, the human immunodeficiency virus (HIV) is the most common to cause infection of the oral cavity. Other RNA viruses causing oral infections include Ebola; enteroviruses causing hand, foot, and mouth disease (HFMD); influenza B and C viruses; and coronaviruses. HIV is an enveloped retrovirus transmitted through sexual contact or by infected body fluids. This virus primarily infects the lymphocytes, resulting in immunodeficiency with consequent clinical manifestations of increased opportunistic infections affecting the lungs and other organs, including the oral mucosa. Interestingly, infectious HIV virions have been shown to be internalized by oral epithelial cells and retained in vesicular compartments for several days. Subsequent contact with lymphocytes or dendritic cells is thought to direct the clinical presentation and potentially promote the viral spread. , Ebola virus is a nonsegmented, negative-stranded RNA virus surrounded by an outer viral envelope studded with viral glycoprotein spikes. The virus primarily causes hemorrhagic fever, and gingival bleeding that is typically associated with other forms of hemorrhage such as epistaxis and bleeding at injection sites. Oral mucosal ulcers, inflammation, and painful red and white patches are other symptoms reported in Ebola virus infections. HFMD is caused by the Picornaviridae family of viruses in the genus enterovirus. Outbreaks of HFMD have more often been caused by two types of enterovirus A species, coxsackievirus A16 (CVA16), or enterovirus 71 (EV71). Oral manifestations are vesicular lesions much like those observed on the skin of hand and feet. ,
Zoonotic Viruses
Influenza and severe acute respiratory syndrome (SARS) heralded the surge of zoonotic virus outbreaks. Globally, influenza epidemics cause major health and economic concerns. These viruses cause self-limiting upper respiratory tract infections in immunocompetent individuals and cause lower respiratory tract infections in immunocompromised subjects and the elderly. Although the primary symptoms are due to respiratory tract infection, the oral cavity could be a major site for the initiation, progression, and pathological processes of the influenza virus–induced infections. Dry mouth, vesiculobullous lesions, aphthous‐like lesions, dysgeusia, and anosmia are common oral manifestations. Coinfection with oral bacteria is thought to aggravate the severity of the lung pathological conditions with increasing morbidity and mortality. ,
Coronaviruses are enveloped, positive-sense, single-stranded RNA (+ssRNA) viruses with a size varying between 26 and 32 kilobases (kb), the largest genome of known RNA viruses. The four subgroups of this family are alpha (α), beta (β), gamma (γ), and delta (δ) coronaviruses. All coronaviruses are zoonotic pathogens, although the mechanisms and routes of transmission from animal to human remain unknown. The primary transmission is thought to be mediated by direct contact with the intermediary host animals, and consumption of milk or uncooked meat have been suggested as potential sources of infection.
This century has seen the emergence of three novel β coronaviruses; namely the SARS-CoV (SARS-CoV-1), the Middle Eastern respiratory syndrome (MERS-CoV), and most recently the novel SARS-CoV-2 that cause severe human diseases and death. The first SARS-CoV-1 outbreak was reported in 2002 in Guangdong, China. The outbreak resulted in more than 8000 infections, with a devastating effect on local and regional economies. The second coronavirus outbreak occurred in 2012 when a few Saudi Arabian nationals were diagnosed with the MERS-CoV infection, which later spread to 27 countries with an approximately 20% to 35% mortality rate.
The third and the most recent outbreak is the infection by the SARS-CoV-2 virus that caused COVID-19. This virus, first reported in the city of Wuhan in China, spread rapidly constituting a global pandemic affecting individuals in all continents. , , As of June 2021, globally over 163 million individuals were infected with SARS-CoV-2 and nearly 3.4 million individuals had died. A critical feature of COVID-19 was the wide variance in clinical severity among infected people and high mortality. In this chapter, we focus on oral manifestations, mechanisms, and significance of oral fluids in SARS-CoV-2 infection.
SARS-CoV-2 and the Oral Cavity
Oral Manifestations of SARS-CoV-2 Infection
SARS-CoV-2–infected individuals present a multitude of oral lesions, including ulcers, petechiae, vesiculobullous lesions, and dysgeusia or ageusia. However, there are considerable inconsistencies in the literature as to the time of oral manifestations with respect to the status of SARS-CoV-2 infection and the stage of COVID-19. Given the large population of SARS-CoV-2–infected individuals globally, some studies have speculated that the oral manifestations could be coincidental or could represent adverse drug-induced responses. Some of the erosive and ulcerative lesions on the oropharynx, palate, and posterior tongue have been suggested as lesions secondary to mechanical intubations. Thus it is not clear whether the oral lesions are true manifestations of the viral infection or a secondary phenomenon resulting from opportunistic infections or adverse effects of treatment.
The rationalr for oral manifestations as true effects of SARS-CoV-2 infection are listed in Box 8.1 . It is well recognized that are SARS-CoV-2 infection is transmitted by aerosols and the oral mucosa, much like the nasopharynx, could be the initial site of contact with the virus. The presence of entry factors for the SARS-CoV-2 in oral tissues suggests susceptibility to infection (discussed later). , , , Further, much of the oral soft tissues are highly vascular and in a state of inflammation supporting increased vulnerability to SARS-CoV-2 infection, as the virus exhibited strong association with vascular inflammation in other organs. , Significantly, a few studies have reported the presence of oral pathogens in the bronchoalveolar lavage fluid from SARS-CoV-2–infected diseased lungs, suggesting coinfection or superinfection.
- •
Much like the nasopharynx, the oral mucosa being exposed to the environment could be the site of first encounter with SARS-CoV-2, and hence oral symptoms could be the first sign of COVID-19.
- •
Inflammatory reactions, in particular vascular inflammation, have been associated with the multiorgan involvement of COVID-19. The highly vascularized oral mucosa is likely to be vulnerable, and the reports of oral ulcers, loss of taste, and petechiae in COVID-19 could be attributed to such inflammatory reactions.
- •
Histopathological similarities were observed between the thrombotic vessels in oral lesions and in the pulmonary diffuse thrombotic disease in COVID-19. ,
- •
Specific entry receptors for SARS-CoV-2 are expressed in oral mucosa.
- •
Oral lesions have been observed in other viral lesions such as the herpes simplex virus, herpes zoster virus, and Epstein-Barr virus infections.
- •
Concurrent or coinfection of SARS-CoV-2 infected lung with oral pathogens have been frequently observed.
Clinical observations during the initial period of the pandemic may not have recorded oral manifestations, particularly in the context of the unexpected severity and mortality associated with COVID-19. As the testing for SARS-CoV-2 infection increased, the emphasis was on identifying early clinical manifestations to prevent and eliminate the rapid spread of infection. Several case reports and a few case series identified variable oral symptoms in SARS-CoV-2–positive individuals, and there was evidence that the oral manifestations could even constitute manifesting clinical features of COVID-19. , ,
In general, taste dysfunction, or dysgeusia, is the most reported oral symptom, with a prevalence rate ranging between 37% and 63% and occasional reports of loss of taste in more than 90% of the case series. , , The initial symptom could be inflamed painful fungiform papillae of short duration of about a day, which then gives rise to an irregular asymptomatic ulcer that often heals completely without scar formation in about a week. , In addition, glossitis and plaque-like lesions on the dorsum of the tongue have been reported in COIVD-19 patients. , , .
A broad classification of oral manifestations of SARS-CoV-2 infection is given in Box 8.2 . Chen et al. first reported xerostomia as a symptom in COVID-19 patients with an equal sex distribution and a prevalence of 46.3% in their study cohort. Subsequent studies also reported xerostomia as an early symptom in asymptomatic health care professionals or mildly symptomatic SARS-CoV-2–positive individuals. The hyposalivation not only results in decreased antimicrobial proteins and peptides but also reduces the efficacy of oral mucosal surface as a physical barrier. In addition, the reduction of saliva secretion may be linked to gustatory dysfunction. , Meta-analysis of available information identified additional oral symptoms such as, aphthous ulcers and vesiculobullous lesions in COVID-19 patients positive individuals. Pooled prevalence of oral manifestations derived from the strength of the observations in the number of infected individuals are listed in Table 8.1 . ( Box 8.3 and Box 8.4 )
- 1.
Chemosensory perception
- a.
Dysgeusia
- b.
Ageusia
- a.
- 2.
Oral mucosal lesions
- a.
Apthous ulcer
- b.
Herpertiform ulcer
- c.
Vesiculobullous lesions
- d.
White lesions, plaques
- e.
Mucositis, mucosal erosions
- a.
- 3.
Salivary gland symptoms
- a.
Xerostomia
- b.
Sialadenitis
- a.
- 4.
Periodontal and bone symptoms
- a.
Bleeding gums, Swelling
- b.
Bone pain
- a.
- 5.
Other:
- a.
Lesions due to trauma associated with intubation
- b.
Drug induced ulcers.
- c.
Secondary opportunistic infections such as oral candidiasis
- a.
| Symptom | Number of Studies (Total No. Patients) | Number of Patients (%) | Reference |
|---|---|---|---|
| Dysgeusia | 40 (10,228) | 10,220 (99%) | |
| 59 (29,349) | 13,615 (46%) | ||
| 140 | 67 (47.9%) | ||
| 111 | 66 (59.5%) | ||
| 1 (573) | 318 (55.5%) | ||
| 3 | 2 (66.6%) | ||
| Oral mucosal lesions | 7 (case reports) | 8 | |
| 140 | 29 (tongue plaque) (20.7%) 6 (lesions on other oral mucosal sites) (4.3%) | ||
| ? (304 nonhospitalized) | 75 (tongue) (24.7%) 3 (other oral mucosal sites) (<1%) | ||
| 74 | 23 (tongue) (31%) 35 (other oral mucosal lesions) (47.3%) | ||
| 1 (573) | 117 (oral ulcers) (20.4%) | ||
| 1 (1237) | 21 (<1%) 3 (tongue) (<1%) 18 (other mucosal regions) (<1%) | ||
| Salivary gland: Xerostomia | 1 (573) | 273 (47.6%) | |
| 140 | 72 (51.4%) | ||
| 111 | 51 (45.9%) |
- 1.
Chronic periodontitis is associated with elevated salivary cytokines and harmful enzymes. Aspiration of salivary cytokines injures the alveolar epithelium, making it more susceptible for viral infection. ,
- 2.
The SARS-CoV-2 spike protein has been shown to interact with the lipid A component of lipopolysaccharide, the cell wall structure of gram-negative bacteria. Because poor oral hygiene is often associated with higher gram-negative content of oral microbiome, preexisting gram-negative bacterial infection, and the associated presence of lipopolysaccharide might exacerbate local lung inflammation. This is a consequence of SARS-CoV-2 spike protein binding by enhanced nuclear factor-κB activation. , This shows that poor oral hygiene is contributing to the increased susceptibility of individuals in nursing care facilities to SARS-CoV-2 infection.
Potential mechanisms by which oral bacteria contribute to the COVID-19 pathogenesis:
- 1.
Oral bacterial migration or transport causing ventilator-associated pneumonia.
- 2.
Oral bacterial infection or superinfection of the lower respiratory tract. ,
- 3.
Oral soft and hard tissue surfaces as reservoirs of respiratory pathogens and as sources of lung reinfection through microaspiration. ,
Advantages
- •
Easy access and feasibility for frequent collection.
- •
Higher compliance to test for SARS-CoV-2 under reduced direct contact.
- •
Positive salivary detection of SARS-CoV-2 in patients with a negative nasopharyngeal swab consistent with true infection.
Limitations
- •
Many studies evaluated saliva admixed with oropharyngeal fluid.
- •
A potential limitation is detection of RNA in levels near the sensitivity limits.
- •
A negative result is not a guarantee of the absence of SARS-CoV-2 infection.
Mechanisms of Oral Manifestations of SARS-CoV-2
SARS-CoV-2 Entry Into Host Cells: A Brief Review
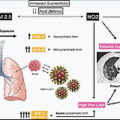
To cause infection, the virus must enter the host cells and replicate. The entry of SARS-CoVs in target cells is facilitated by the interaction between the spike proteins protruding from the surface of the virus and the host cell surface receptors. In silico, molecular and functional assays have identified a metallopeptidase cell surface receptor named angiotensin-converting enzyme-2 (ACE2) on host cells as a common entry receptor for all coronaviruses. The spike (S) protein of SARS-CoV-2 is composed of two subunits, S1 and S2. The S1 subunit contains a receptor-binding domain that recognizes and binds to the host receptor ACE2, whereas the S2 subunit mediates viral cell membrane fusion by forming a six-helical bundle by the two-heptad repeat domain. SARS-CoV-2 has been shown to bind ACE2 with higher affinity than SARS-CoV-1. Other host cell proteases that facilitate or promote the viral entry include furin and the transmembrane protease serine (TMPRSS). Furin, ACE2, desintegrin, and metalloproteinase domain 17 (ADAM17) or tumor necrosis factor-alpha (TNF-α)–converting enzyme (TACE) are proteases that cleave the viral spike protein between S1 and S2. Whereas the S1 domain potentially facilitates receptor binding, the S2 domain regulates the fusion of the virus with the host cell. , The TMPRSS belongs to a family of 17 members, divided into four subfamilies (HAT/DESC, epsin/TMPRSS, matriptase, and Corin). The TMPRSS2 of the epsin subfamily has been shown to participate in mediating SARS-CoV-2 entry. Much like furin, the TMPRSS2 also cleaves the spike protein between the S1 and the S2 subunits. The S2 subunit then undergoes conformational changes to complete the fusion of the viral envelope with the host cell membrane, mediating receptor internalization and facilitating viral entry. Replication within the target cells establishes the SARS-CoV-2 infection, leading to subsequent tissue damage. ,
Pathogenesis of Oral Mucosal Lesions in COVID-19
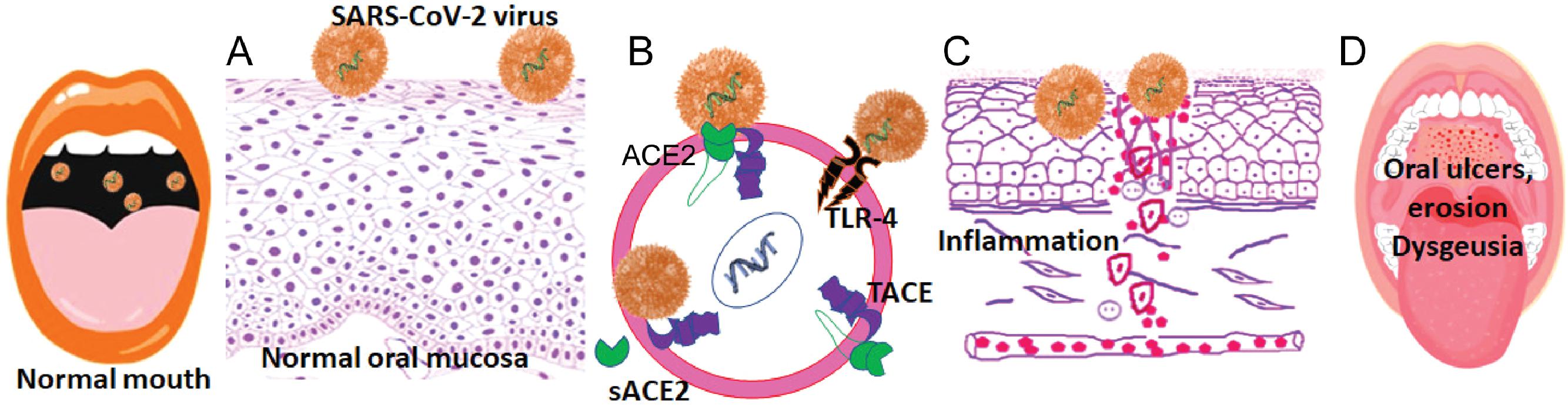
In humans, ACE2 is predominantly observed in epithelial cells in contact with the environment, although expression also has been reported in internal organs, including the kidney and the heart. , , The oral cavity in direct contact with the external environment is at an increased risk for viral invasion. Although one of the primary functions of the oral mucosa is to provide a barrier against microbial invasion, the surface epithelial cells often express receptors that would allow attachment and penetration of the cell membrane by the microbes. Based on the analyses of two public RNA sequence databases, Xu et al. first reported that ACE2 was differentially expressed in the oral mucosa in different sites, with the highest expression in the tongue and floor of the mouth. They confirmed the high expression of ACE2 in oral epithelial cells by single cell transcriptome analysis of normal oral mucosal biopsy samples. Subsequently, we and others also showed that ACE2 is abundantly expressed in the lining mucosa of the oral cavity and the dorsum of the tongue ( Fig. 8.1 ). In addition, the oral epithelial cells also express furin and TMPRSS2, supporting the potential for viral spike protein cleavage and SARS-CoV-2 internalization. The expression of TMPRSS2 is also observed in the acinar, myoepithelial and the epithelial cells lining the ducts in the salivary glands. , , SARS-CoV-2 transcripts have been identified in oral mucosal tissue autopsies from COVID-19 patients.
Emerging evidence suggests that in addition to ACE2, the SARS-CoV-2 also uses other pattern recognition receptors such as Toll-like receptors (TLRs) for host cell entry (see Fig. 8.1B ). In particular, the observations of TLR upregulation by damage-associated molecular patterns in clinical samples and the proposed models of direct binding of spike protein with TLRs 1, 4, and 6 suggest specific roles for these TLRs in facilitating the viral entry and/or in mediating the clinical symptoms of COVID-19. It has been suggested that the dysbiosis secondary to the invading SARS-CoV-2 increases the prevalence of pathogenic bacteria, which in turn modulates the innate host responses resulting in the release of cytokines and inflammation. , Such inflammatory reactions secondary to viral entry in mucosal and vascular cells has been used to explain the oral erosion, ulcers, and lingual pain in COVID-19. Collectively, these observations suggest that SARS-CoV-2 can infect a wide variety of oral tissues and cells and provide the scientific rationale for the oral symptoms in infected individuals.
Pathogenesis of Hyposalivation in COVID-19
SARS-CoV-2 Entry Factors in Salivary Gland Tissues
Xerostomia is a common complaint in COVID-19, suggesting that the virus potentially infects the salivary glands. Substantiting the clinical symptom are the molecular characteristics of oral tissues exhibiting abundant expression of ACE2, the host cell receptor of SARS-CoV-2 entry, in the parotid gland acinar cells and in the epithelial cells lining the salivary gland ducts as determined by single-cell RNA-sequencing atlases and clustering analyses. A high expression of ACE2 has been reported in the granular cells and the ductal epithelial cells of the salivary glands. , Further, genotype‐tissue expression studies have shown that TMPRSS2 is moderately expressed in the salivary glands. In particular, the relative expressions of ACE2 and TMPRSS2 in the salivary glands was similar to that in the respiratory and GI epithelia that are prone to SARS-CoV-2 infection. The presence of ACE2 and TMPRSS2 at both the genetic and the protein levels have been futher confirmed by in situ hybridization and immunohistochemistry. , A high expression of ACE2 has been reported in the granular cells and the ductal epithelial cells of the salivary glands. ,
SARS-CoV-2 Infection and Replication in Salivary Gland Tissues
SARS-CoV-2 transcripts have been detected in postmortem salivary gland tissues of COVID-19 patients. , The viral RNA was detected by quantitative PCR (qPCR) in the parotid and submandibular salivary gland tissues and in the minor salivary gland tissues. Histological examination showed that the ductal epithelial cells of the salivary glands exhibited morphological changes consistent with viral infection, including cytoplasmic and nuclear vacuolization and nuclear polymorphism. The glandular acinar cells exhibited enlarged nuclei and degenerating zymogen granules. Electron microscopy showed that the acinal cells and the epithelial cells lining the salivary gland ducts exhibited spherical 70- to 100-nm viral particles, consistent in size and shape with the Coronaviridae family. These cells also exhibited degeneration of organelles in infected cells and the presence of a cluster of nucleocapsids suggestive of viral replication. , Interestingly, the SARS-CoV-2 viral load was observed to be higher in minor salivary gland tissues as compared to that in the parotid tissues. In situ hybridization and immunohistochemistry showed increased presence of the viral transcripts in the minor gland ductal epithelial cells and in the acinar cells expressing ACE2, although significant variability was observed in the extent of individual cell infection. Significantly, these observations in postmortem salivary gland tissues were also observed in minor salivary gland tissues from acutely infected individuals with COVID-19. ,
Collectively, based on the expression and distribution of SARS-CoV-2 entry factors in the salivary gland tissues and the potential infectivity, it is suggested that the SARS-CoV-2 enters the epithelial cells and replicates, causing acute sialadenitis and destruction of acinar cells. Resolution of the acinar cell inflammation by fibroblast proliferation and connective tissue fibrosis, could lead to hyposalivation and the consequent symptom of dry mouth. Sialadenitis and dry mouth are frequently reported as common symptoms of COVID-19. , , ,
Pathogenesis of Loss of Taste in COVID-19
As discussed earlier, systematic analyses showed that the gustatory dysfunction is one of the most common symptoms of COVID-19. , , , , The epithelial cells of the dorsum of the tongue, including the taste bud cells, have been shown to coexpress the viral entry receptors ACE2 and TMPRSS2. , , Indeed, the density of ACE2 receptor expression has been observed to be higher in tongue tissues than that in the buccal mucosa or gingiva. , , Sakaguchi et al. showed by immunohistochemisry that ACE2 and TMPRSS2 are localized in human fungiform papillae taste cells. Significantly, Han et al. showed by single cell profile analysis of tongue tissues that the distribution of ACE2-positive cells correlated with typical the gene markers of taste buds, including TAS1R3, TAS2R4, TAS2R14, SNAP25, and NCAM1. Furthermore, taste bud cells have been shown to express TLRs more abundantly than the nongustatory lingual epithelium. Specifically, TLRs-2, -3, and -4 are highly observed in the gustducin-expressing type II taste bud cells. , Thus the expression of multiple viral entry receptors makes taste bud cells highly susceptible for SARS-CoV-2 infections. , Pertinently, human taste bud cells have also been shown to support SARS-CoV-2 replication. Biopsies of the dorsum of the tongue from COVID-19 patients showed transcripts of the SARS-CoV-2 by real-time polymerase chain reaction (PCR). Direct infection of the taste bud cells promotes inflammation and this could affect taste perception. , , Consistently, gustatory dysfunctions have been correlated with high serum interleukin-6 (IL-6), a key cytokine upregulated in acute and persistent SARS-CoV-2 infection.
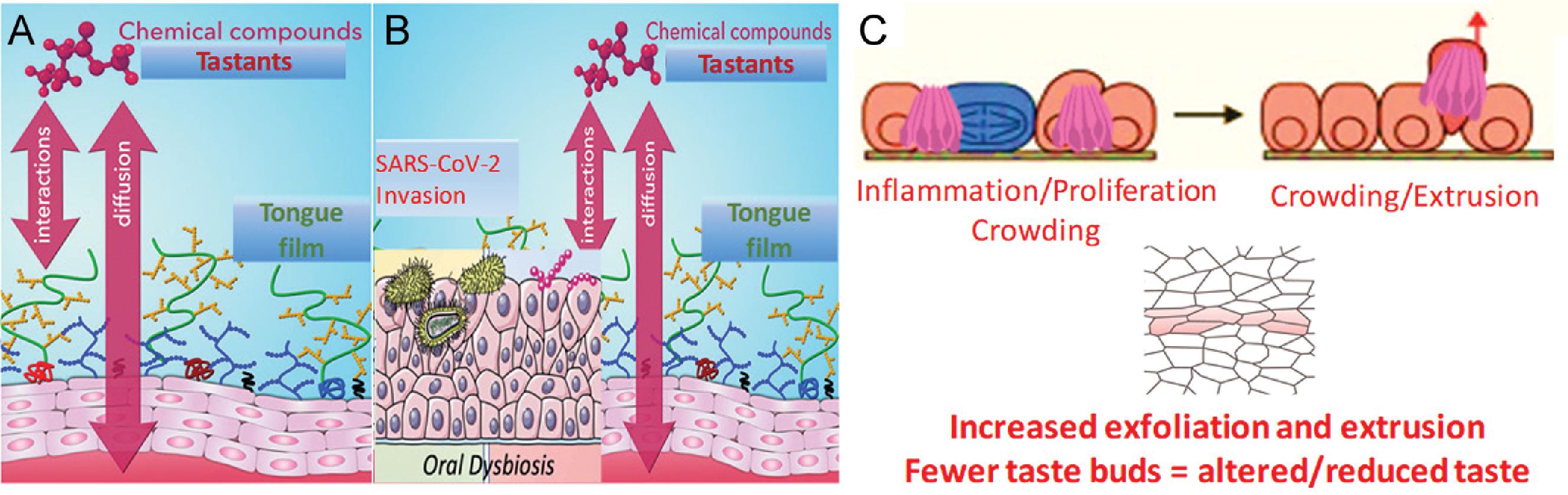
Taken together, a conceptual framework for the pathogenesis of dysgeusia in long-COVID could be proposed ( Fig. 8.2 ). SARS-COV-2 infection of the tongue epithelial cells, including the taste buds, modulates the host responses, and prolonged exposure will precipitate an exaggerated inflammatory reaction. Mucosal inflammation increases the epithelial cell exfoliation, a source of viral shedding in saliva and a potential cause for invasion of bystander epithelial cells. The lag in replenishment of the lost cells together with the reduced stem cell turnover could result in fewer taste buds. The reduced sensory perception precipitate dysgeusia. It will be interesting to investigate whether salivary epithelial cell analyses could reveal specific markers of dysgeusia in individuals with long-COVID.
The Oral Microbiome and COVID-19
The Oral Flora and Lung Microbiome in Health
A healthy human inhales 12 to 15 times per minute at rest, on average, with approximately 20% of the air in the lungs being exchanged during each breath. The popular belief that the healthy lungs are sterile has been dismantled by a large body of evidence that supports the presence of a healthy lung microbiome, which when disturbed can contribute to many lung pathological conditions. Multiple theories have been proposed for the development of the lung microbiome. , The logical topological community theory proposes that the initial seeding of the lung microbiome occurs from the nearby anatomical sites based on the species similarity between the oropharyngeal and the nasopharyngeal microbiome with that of the lung microbiome. , The proximity and the anatomical continuity of the oral cavity and the lower respiratory tract suggests that the oropharyngeal microbiota could be a major contributor to the lung microbiome. Indeed, compositionally, bacterial communities from the lung are observed to be indistinguishable from the upper airways, albeit about 2 to 4 logs lower in biomass. The air passing through the oropharynx is admixed with saliva laden with inhaled microbes, oral microbes, and trapped particulates. The movement of the air not only assists in the development of the aerodigestive mucosal biofilm but also plays a role in the frequent modulation of the microbiome secondary to the flow and the composition of salivary microbiota. , Further movement of the epiglottis that diverts the solid contents to the esophagus and the air to the respiratory tract creates biologically significant number of aerosols. Microaspiration of these aerosols contributes to the vast majority of the constant microbial seeding of the lower airways from the oral cavity during health. , , Thus, in general, the healthy lung does not contain a constant or distinct microbiome, but instead contains low levels of bacterial sequences largely similar to those of the upper respiratory tract flora. , Analysis of 16S rDNA and deep sequencing suggest that the lung-specific sequences are not shared among individuals. The biogeography theory posits that the spatiotemporal variability of the lung is influenced by the continual stream of immigrant bacterial species by microaspiration and selective replication of specific bacteria taxa. , Hence, this theory provides a rational explanation for the observations that despite the close resemblance of the lung microbiome to the oral microbiome, certain bacteria are represented in higher abundance in the lung microbiome than expected from the corresponding oral microbiomes. This is likely due to a selective advantage in replication among those bacteria taxa in the lung microenvironment as opposed to the oral microenvironment.
The Role of Oral Bacteria in Lung Microbiome Dysbiosis
As stated previously, the balance of three factors determines and shapes the lung microbiome: (1) microbial immigration into the airways, (2) elimination of microbes from the airways, and (3) the relative reproduction rates of the microbes found in the airways as influenced by the local environment for microbial growth. Acute and chronic lung diseases can dramatically change the ecological determinants of the lung microbiome, resulting in markedly different microbial communities. Inflammation alters the lung microbiome by affecting the regional growth conditions as a result of changes imposed in nutrient availability, temperature, pH, and oxygen tension. , A shift in the microbiome composition away from the Prevotella, Veillonella, and Streptococcus spp. that dominate the healthy lung microbiome toward Gammaproteobacteria, the class that contains many common gram-negative “pathogens” associated with lung diseases, has been observed in inflammatory pathological lung conditions.
Epidemiological studies support the notion that the oral flora is of relevance to acute and chronic lung disease. , , An increase in bacterial immigration from the oropharynx occurs, and several oral anaerobes and facultative species have been cultured from infected lung fluids. These include Porphyromonas gingivalis, Bacteroides gracilis, Bacteroides oralis, Bacteroides buccae, Eikenella corrodens, Fusobacterium nucleatum, Fusobacterium necrophorum, Actinobacillus actinomycetemcomitans, peptostreptococci , Clostridium, and Actinomyces. , , Although poor oral health has been associated with an increased risk for respiratory disease, improving oral health resulted in a reduction of respiratory events among patients in nursing homes and intensive care units (ICUs). ,
Several mechanisms have been postulated for the role of oral bacteria in the pathogenesis of respiratory infection. Aspiration of oral pathogens into the lower respiratory tract can precipitate lung infection. Cytokines originating from periodontal tissues may alter respiratory epithelium promoting invasion and infection by bacterial pathogens. , Alternatively, periodontal disease–associated enzymes in saliva may modify the mucosal surfaces to promote adhesion and colonization by respiratory pathogens. Furthermore, backward mucus flow by the ciliary epithelium of the bronchial mucosa toward the oropharynx permit bidirectional movement of the microbiome during the coughing reflex. , , The oral mucosa thus could act as a reservoir of respiratory pathogens, which are then aspirated into the lung with salivary droplets. , ,
Coronavirus Infections, Bacterial Coinfections, Pathological Lung Conditions, and the Role of the Oral Microbiome
The Triad of Coronavirus Infections, Bacterial Coinfections, and Pathological Lung Conditions
Considerable evidence suggests that the influenza and other respiratory viral infections predispose patients to secondary bacterial coinfection or superinfections, often leading to severe clinical disease. During the 1918 Spanish flu pandemic by H1N1 influenza virus, secondary pneumonia resulting from bacterial superinfection had a major role in the high mortality rates experienced globally. High mortality secondary to bacterial coinfection was also detected in approximately 30% of cases in the 2009 H1N1 pandemic despite administration of appropriate antibiotics. ,
Clinical symptoms of COVID-19 infections range from mild or moderate flu-like symptoms to severe pneumonia requiring oxygen support. , Meta-analysis, systematic reviews, and clinical data show that the predominant causes of mortality in COVID-19 are pathological conditions of the respiratory system, including pneumonia, chronic lower respiratory tract disease, respiratory distress syndrome, respiratory failure, and multiorgan failure secondary to suppurative pulmonary infection. Many of these symptoms and pathological conditions in COVID-19 patients are attributed to bacterial or fungal coinfections and are associated with poor clinical outcome, especially in critically ill patients. , ,
Interestingly, it has been suggested that as opposed to two other beta coronaviruses, SARS-CoV-1 and MERS-CoV, the SARS-CoV-2 infection facilitated/promoted increased secondary bacterial and/or fungal coinfections or superinfections. The difficulty in obtaining site-specific samples without cross-contamination with oropharynx and saliva is a hurdle in characterizing the upper and lower respiratory tract microbiome. However, few available reports on the analysis of bronchoalveolar lavage samples suggest that the lower respiratory tract or the lung microbiota of deceased COVID-19 patients included complex colonization by bacterial and fungal opportunistic species. , Although the alpha diversity of the lung microbiome did not differ between the COVID-19–positive and COVID-19–negative patients, significant changes in the microbial composition and relative abundance of specific bacteria have been observed in COVID-19 lung tissues as compared with that in the healthy lungs. Indeed, investigating the lung and the blood microbiome of COVID-19 patients by high-throughput RNA sequencing, Dereschuk et al. observed that 91 bacterial and 14 fungal species were differentially abundant in lung biopsy samples, 13 bacteria and 9 fungal species were differentially abundant in the blood mononuclear cells, and 12 bacteria and 57 fungal species were differentially abundant in bronchoalveolar lavage fluid.
The Role of Oral Microbiome in COVID-19 Lung Disease
The mechanisms of bacterial coinfection or superinfection during or after viral infections consist of complex multifactorial processes mediated by interactions among the viruses, the bacteria, and the host immune system ( Box 8.3 ). Direct mucosal/epithelial damage by the virus could modulate microbial colonization, causing dysbiosis of the respiratory mucosa, and the consequent dysregulations of the immune responses increase the susceptibility to secondary bacterial coinfections. Alternatively, bacterial pneumonia can develop secondary to aspiration of pathogenic oral/oropharyngeal bacteria into the lower respiratory tract. , , Pertinently, the Bacteroides spp. identified as abundant in the lung tissues, in the bronchoalveolar lavage fluid, and the blood of COVID-19 patients are common colonizers of oral flora. At the phylum level, the Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria reported in COVID-19 are also abundant in the normal oral microbiome. , Interestingly, a similar specific oral bacterial preponderance that influenced pneumonia development was previously reported in influenza infections. Although these studies have not correlated the lung microbiome with the oral flora, the observations support translocation of oral bacteria into the respiratory tract, with aspiration of saliva the primary mode. The salivary microbiota is a “conglomerate of bacteria shed from oral surfaces” and could serve as a reservoir of multiple pathogens. Significantly, it has been suggested that not only the microbial translocation but also that of bacterial components from saliva could contribute to the disease aggravation in COVID-19. The SARS-CoV-2 spike protein has been shown to bind the lipopolysaccharide (LPS), and the interaction is mediated between the lipid A component of the LPS and the S1/S2 furin cleavage site in the spike protein. In silico docking studies showed that the interaction of LPS with the spike protein is structurally very similar to the binding orientation of LPS with its coreceptors, CD14 or MD2. Functionally, the LPS potentiated the proinflammatory effects of the SARS-CoV-2 spike protein, as shown by upregulated nuclear factor-kappa B (NF-κB) activity in cells exposed to both LPS and the spike protein. , Pertinently, mass spectrometric characterization of salivary LPS suggested that the saliva of patients with chronic periodontitis possessed hyperacylated and phosphorylated lipid A isoforms that are highly immunogenic. In this context, it is tempting to speculate that the aspiration of saliva with the chemically active form of LPS could potentiate further the inflammatory responses of SARS-CoV-2 infection and aggravate the lung pathological condition.
As the name suggests, severe acute respiratory distress is the most common cause for hospitalization in COVID-19. , Hypoxemia is common in hospitalized patients, and mechanical ventilation is the mainstay of management. Ventilator-associated pneumonia remains a frequent serious complication of these treatment modalities and is often associated with increased mortality. Pathologically, the ventilator-associated pneumonia has been attributed to the dysbiosis in the lower respiratory tract. Microbial modifications in the oropharyngeal regions and bacterial migration to the respiratory tract has been suggested as the nidus for transcolonization and the dysbiosis. The source of the microbial shift in oropharynx could be from either the gut flora or the respiratory tract. It is suggested that the oral epithelium provides niche surfaces for adherence of the bacteria from the respiratory tract that potentially act as reservoirs. , Retranslocation to the respiratory tract by microaspiration is suggested as yet another cause for ventilator-associated pneumonia. Immunoinflammatory responses to the altered microflora leads to pneumonia, tissue loss, and consequent death. ,
The Role of Oral Microbiome in Causing Taste Dysfunction in COVID-19
As stated earlier, the lingual epithelial cells, including the taste receptor cells, express multiple entry factors for SARS-CoV-2 virus. Indeed, the SARS-CoV-2 has been shown to replicate in human taste bud cells supporting infective potential. Furthermore, the SARS-CoV-2 invasion could promote a favorable environment for coinfections and contribue to the variable symptoms of ageusia or dysgeusia that either exist for short duration or persist for longer periods. Analyzing oropharyngeal swabs from hospitalized COVID-19 patients, Iebba et al. reported that a select panel of oral bacteria and cytokines is predictive of neurological symptoms, including hyposmia and dysgeusia in SARS-CoV-2–infected individuals. Specifically, the bacterial genera of Streptococcus, Veillonella, Prevotella, Lactobacillus, Capnocytophaga, Porphyromonas, Abiotrophia, Aggregatibacter, and Atopobium were increased in swabs from COVID-19 patients compared to that from with healthy (SARS-CoV-2 non infected) controls, whereas the genera of Rothia, Haemophilus, Parvimonas, Fusobacterium, and Gemella were decreased in oral samples in COVID-19 patients. , In this context it is interesting to note that a previous study reported that the relative abundance of the Fusobacterium and Rothia on the dorsum of the tongue correlated positively with the perception of sweet and salt taste, respectively. However, only few studies have addressed the role of the oral microbiome in taste perception. Although it is tempting to speculate that the changes in the bacterial genera observed in COVID-19 patients could be at least partially related to the symptom of dysgeusia, more data are needed to resolve the pathogenesis of the condition. Furthermore, in COVID-19, it is not known whether oral dysbiosis is the cause or effect of SARS-CoV-2 infection and whether it could be related to postviral complications.
Salivary Diagnostics, Transmission, and Biomarkers for COVID-19
Saliva for Diagnosis of SARS-CoV-2 Infection
The COVID-19 diagnosis is based on the detection of SARS-CoV-2 RNA by real-time PCR. Molecular diagnostic tests for quantitating the viral RNA include the traditional bench real-time qPCR system, a high-throughput real-time qPCR system (automated from RNA extraction to reporting of results), and direct rapid RNA extraction-free real-time qPCR kits have been widely used globally (3). Additional methods include reverse transcription–loop-mediated isothermal amplification and a rapid antigen test that combines immunochromatography with an enzyme immunoassay to detect the viral nucleocapsid (N) protein, for potential use as point-of-care tests.
Although SARS-CoV-2 RNA detection in the nasopharyngeal swab is considered the gold standard method for COVID-19 diagnosis, demand for swabs early in the pandemic drove a rapid collapse of supply chains, with shortages of personal protective equipment required by health care workers for sample collection. Further, the sample acquisition involved close contact of health care workers with the infected or suspected SARS-COV-2–positive individuals, increasing the risk for transmission. In addition, the specimen collection procedure was invasive and uncomfortable for patients. These shortcomings fostered search for use of alternative sampling methods for detecting the virus. More importantly, the rapid spread of the virus necessitated urgent development of methods for repeated sampling and mass testing.
Alternative samples assessed for SARS-CoV-2 detection include pooled nasal mid-turbinate swabs, throat swabs, oropharyngeal wash, and saliva. Each of the sample collection methods and the validity of the specimen with respect to SARS-CoV-2 detection has merits and drawbacks. Although all alternative methods were attempted as self-collected specimens with no contact with health care workers to minimize the risk for transmission, saliva collection was easier, less invasive, and more acceptable. Mishra et al. conducted a retrospective analysis of the validity of nonrespiratory samples for detecting the coronaviruses responsible for the previous three pandemics of the 21st century. Assessing data from a total of 3274 that included 802 SARS-CoV-1 samples and 2347 SARS-CoV-2 samples, they found that the sensitivity of nonrespiratory specimens for detection of SARS-CoV-2 was lower at 57.5% (95% CI, −1.2%–116.2%) than that for SARS-CoV-1 at 96.7% (95% CI, 87.6%–100.0%).
Previously, during the SARS-CoV-1 pandemic, a high concordance was observed between the saliva and the nasopharyngeal swabs as diagnostic specimens. Use of saliva for detection of SARS-CoV-2 was first reported by To et al., during the initial outbreak. They assessed the saliva of 12 nasopharyngeal swab–confirmed COVID-19 patients between 0 and 7 days of hospitalization. They reported a median viral load in saliva at 3.3 × 10 6 copies/mL, with a range of 9.9 × 10 2 and 1.2 × 10 8 copies/mL. , Subsequent studies testing saliva alone or testing both saliva and paired nasopharyngeal swab samples suggested that the SARS-CoV-2 titer in saliva was equivalent or even higher than that in the nasopharyngeal swab. The sensitivity of SARS-CoV-2 RNA detection in saliva ranged between 81.6% and 100%, and the specificity ranged between 88.4% and 100%. , , Indeed, the virus was detected in the saliva of asymptomatic individuals, albeit at lower titers (10 4 –10 5 copies/mL) when the nasopharyngeal swabs were negative. Because the real-time PCR is based on well-characterized and validated primers, detection of SARS-CoV-2 RNA in saliva is not classified as false positive but rather as false negative or misclassification of nasopharyngeal samples. Further, in contrast to the inconsistent nasopharyngeal swab results, the viral titer in saliva decreased progressively in severely ill COVID-19 patients during recovery. In this context, the temporal analysis of SARS-CoV-2 viral load in saliva could serve a pivotal role for the translation of salivary tests in the clinic. Furthermore, glandular saliva from the parotid gland duct yielded high titers of the virus suggesting that SARS-CoV-2 targets the ductal and/or glandular epithelial cells. , Interestingly, fecal-oral transmission has been suggested as a source of the virus in saliva in COVID-19 patients presenting with GI symptoms with fecal shedding of SARS-CoV-2. ,
Encouraged by the data from early studies, the use of saliva as a specimen for detecting SARS-CoV-2 increased globally. Several systematic reviews and metanalytic studies conducted comparative analyses of the sensitivity of saliva and that of the gold standard nasopharyngeal swabs as biospecimens for detecting SARS-CoV-2 , , , ( Table 8.2 ). Collectively, there is evidence that the saliva as specimen exhibits high sensitivity and specificity for detecting SARS-CoV-2 ( Box 8.4 ). Further, the feasibility of frequent and self-collection of saliva are distinct advantages in infections with rapid spread and transmission such as SARS-CoV-2. Indeed, Eduardo evaluated 55 self-collected unstimulated saliva samples from hospitalized COVID-19 patients and demonstrated that the saliva samples were 87.3% in agreement with the nasopharyngeal swab for the detection of SARS-CoV-2 virus. In contrast, in studies comparing paired nasopharyngeal swabs and self-collected saliva obtained simultaneously, saliva samples exhibited lower sensitivity ranging from 16% to of 24.4%. Nagura-Ikeda et al. reported that the viral RNA was detected at significantly higher percentages (65.6%–93.4%) in saliva specimens self-collected within 9 days of symptom onset. However, the virus detection was lower as the time passed after at least 10 days of symptoms (22.2%–66.7%) and in specimens collected from asymptomatic patients (40.0%–66.7%). 139 In asymptomatic individuals, random saliva samples (92.3%) were superior or equivalent to the pooled nasopharyngeal and oropharyngeal samples (73.2%) for detecting the SARS-CoV-2 virus by amplification of the open reading frame 1a (ORF1a) and nucleocapsid (N) genes. Reanalysis of SARS-CoV-2–positive frozen saliva samples by immunochromatographic assay and chemiluminescent enzyme immunoassay detected SARS-CoV-2 in 41% by the former method and in 91% of samples by the latter method.