Nexgen pathology: predicting clinical course and targeting disease causation
Carlos Cordon-Cardo, MD, PhD  Adolfo Firpo-Betancourt, MD, MPA
Adolfo Firpo-Betancourt, MD, MPA
Overview
Pathology is a bridging discipline involving basic and clinical biomedical sciences. This context includes both descriptive and mechanistic approaches, with the goals of understanding the anatomical changes and underlying molecular events involved in disease-related processes. Neoplastic disorders are a focal point for the further development of this chapter. The two main objectives of pathology are to define disease causation [from “Pathos” (Greek) “Disease”] and categorize disease states to render clinical diagnostic services. A modern academic Department of Pathology encompasses education activities, including teaching students, training residents and fellows, and mentoring faculty; and hospital-based clinical services usually under three divisions, comprising anatomic pathology (surgical pathology, cytology, and autopsy services); clinical pathology (embodying a variety of laboratory services from blood bank and coagulation to chemistry and microbiology, among others); and molecular pathology (commonly housing somatic genetics, cytogenomics, and flow cytometry). During the past two decades, we have witnessed the transition from descriptive analysis of tissue histology and analyte variables that categorized patients and broad disease stages to more objective and quantitative multidimensional studies aimed at defining individual patient signatures. More traditional population and cohort-based classifications are turning into patient-specific profiles that optimize treatment efficacy and outcome: from diagnostic and prognostic approaches that group patients into disease categories to the development of a more precise, predictive, and individualized patient assessment (Table 1). Such an integrated care model drives selection of evidence-based treatment protocols to optimize clinical outcome, engendering a cost–effective and personalized healthcare. Disease classification and assistance in selection of therapy is the focus of this “patient-centric” pathology approach, expanding into monitoring of therapy (such as assessing therapeutic index and mutational load through tumor genotypes) and managing high–risk patients through early diagnosis of their disease condition (Figure 1). The ultimate goals are to move from treating symptomatology to treating disease causation once origin of the disease is better understood and to move from a fee-for-service to population-based accountable healthcare management.
From anatomic and clinical pathology to molecular and predictive integrated diagnostics
The origins and the academic establishment of pathology can be traced to the sixteenth to seventeenth centuries in Italy, where physicians systematically performed autopsies to further elucidate diseases suffered by their patients. It is in the writings of Benivieni (1443–1502) and Morgagni (1682–1771) that we find the first clinicopathological correlations, based on meticulous autopsy studies. This descriptive, gross anatomic approach was supplanted by the French and English “tissue pathology” schools, represented by Bichat (1771–1802) and eventually by the German school of “cellular pathology,” led by Muller (1801–1858) and Virchow (1821–1902).1 The cell was defined as the unit of life and the target for most diseases, being further expressed by histopathological alterations in the tissue of origin (Figure 2).2 Biochemical, immunological, and genetic discoveries brought scientific knowledge into clinical medicine, and a new armamentarium of technologies during the mid-to-late 1900s. An upsurge of innovative approaches, from DNA amplification and mutation-identification platforms to automated clinical chemistry appeared in the late twentieth century. Molecular pathology evolved from these investigations, integrating histology with immunohistochemical (IHC) and genetic technologies to render phenotype/genotype correlations, thus establishing individualized patient assessment (Figure 3).3, 4
Table 1 Evolution of pathology from diagnostic to integrated predictive platforms
| Classical pathology | Molecular and systems pathology | |
| Diagnosis and staging | Descriptive analysis of clinical variables, tissue histology, and biomarkers to categorize patients into broad disease stages; no predictive value to inform treatment selection | Objective, quantitative multidimensional analysis of clinical variables, tissue morphometrics, and molecular tumor signatures to define individual patient tumor phenotype and genotype |
| Prognostic evaluation | Traditional population and cohort-based classification used to deduce disease progression and likelihood of treatment response; nonspecific | Patient-specific characteristics and molecular tumor profiles are used to predict drug sensitivity and radiation response, thus optimizing treatment efficacy and outcome |
| Treatment selection | Group management approach that stratifies patients into disease categories, assigning therapies on predetermined population-based protocols instead of being patient specific | Personalized and integrated care model drives selection of evidence-based treatment protocol to optimize clinical outcome; patient-tailored treatments improve survival and quality of life |
| Diagnostic and prognostic approach that “groups” patients into disease categories | Precise, predictive, and cost effective; individualized patient management |
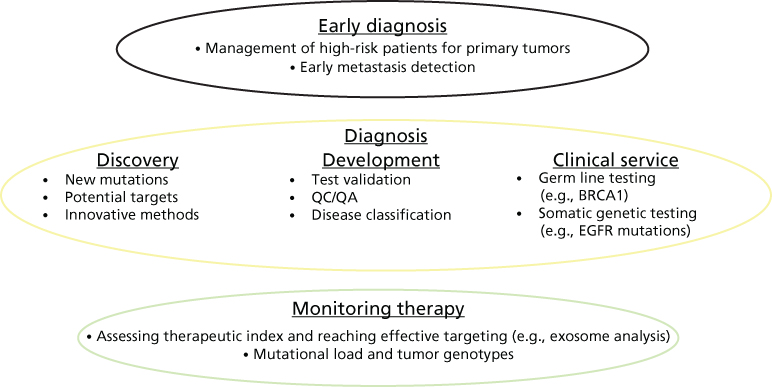
Figure 1 Integrated approach to disease management: translating data into knowledge.
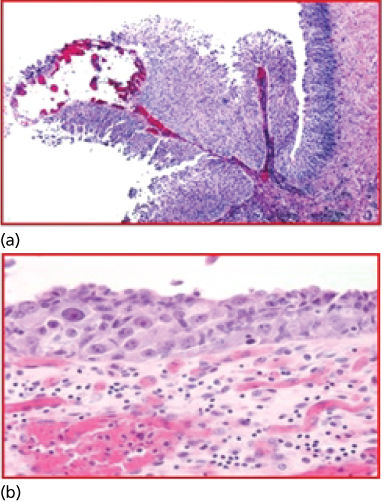
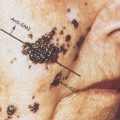
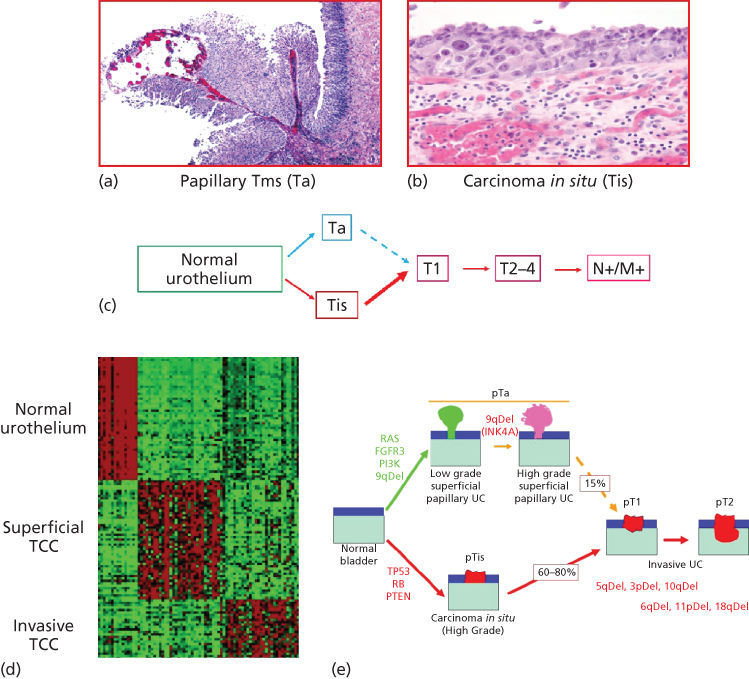
Figure 2 (a, b) Cellular profiling. The microscope became the innovative device that allowed Dr. Rudolf Virchow (1821–1902) to describe the cell as the unit of life and main target of disease. Disease classification based on histogenesis was developed. Hematoxylin and eosin (H&E) of bladder cancer histological sections; upper microphotograph illustrating a papillary noninvasive transitional cell carcinoma; lower microphotograph illustrating a transitional cell carcinoma in situ of the urinary bladder.
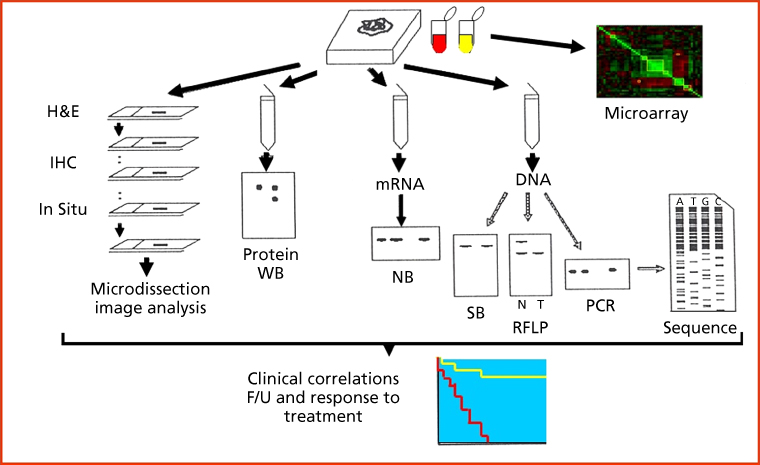
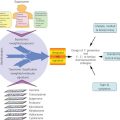
Figure 3 Molecular pathology. Integration of histological, immunochemical, and genetic approaches defines the nature of molecular profiling. Phenotype and genotype are integrated using low- and high-throughput technologies, including histology, protein studies, and nucleic acid analyses. The use of microarray technologies allows the interrogation of multiple analytes on a single assay. H&E: hematoxylin and eosin stain; IHC: immunohistochemistry; WB: western blot; NB: northern blot; SB: southern blot; PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism; F/U: follow-up.
The role of the pathologist has changed over the years. Traditionally, the main task of the pathologist was the evaluation of patients through morphologic analyses of human tissues and analysis of certain chemicals and biomolecules in body fluids. The diagnosis of cancer involves the analysis of tissue samples and body fluids for features that correlate with malignant transformation and tumor progression. Tissue specimens, cells, and fluids (e.g., blood, urine, and effusions) are obtained through several procedures, including surgical biopsy, endoscopic biopsy, core or aspirational needle biopsy, venipuncture, spinal tap, scraping of tissue surfaces, and collection of exfoliative cells from urine or sputum. The acquired tissue or cell specimens are subjected to a series of analytic modalities for diagnostic purposes. Light microscopy, assessing morphological features of the procured specimens, was utilized almost as the sole approach for many years and remains the standard diagnostic method to which all novel methods need to be compared. The use of enzyme histochemistry and electron microscopy expanded the primary microanatomical evaluation to include biochemical and subcellular structural features. More recently, cytogenetics, analysis of DNA content, molecular genetic assays, and IHC studies have been added as valuable adjuncts to light microscopy in cancer diagnosis, expanding to an in-depth genotype–phenotype characterization. These methods, particularly IHC, have enhanced significantly our ability to define the lines of differentiation of human tumors. The inclusion of these technologies with more recent high throughput, such as next generation DNA sequencing platforms and RNA expression arrays among others, constitutes the basis for molecular pathology, and has added valuable information to the current classification schemes.5 The main goal now is to integrate the data that emanate from this multidimensional arsenal of technologies and translate it into knowledge and to manage such knowledge in order to better manage our patients and community of individuals at high risk for developing certain diseases.
Molecular pathology
Molecular pathology studies the origins and pathogenesis of disease at the biochemical, immunological, and genetic levels. Cancer is a clonal disease driven primarily by genetic mutations and epigenetic derangements.6, 7 This means that any given tumor originates in a single cell (clonality) that has been primed by specific molecular triggers (tumorigenesis). Such a cell accepts genetic manipulation, does not activate pathways of cell death or immune surveillance, and progresses toward invasion and metastatic spread, the ultimate malignant phenotype. Even though several tumors have their origin in viral infections (oncoviruses) in which certain viral proteins (oncoproteins) enhance proliferation and abolish apoptotic responses, such as the case of certain human papilloma virus subtypes, most cancers can be defined as diseases driven by either inherited or somatic mutations. In this regard, pathology differs from medical genetics, in which the main objective is to determine the abnormalities associated with inherited disorders carried as germ-line mutations. Molecular pathology of cancer aims to identify and understand the aberrations involved in the development and progression of neoplastic diseases. Clinically relevant objectives include (1) establishing a definitive diagnosis and classification of tumors, based on the recognition of complex profiles (fingerprints) or unique molecular alterations that occur in specific tumor types; (2) providing early detection of tumor cells using sensitive molecular techniques, thus anticipating therapeutic intervention; (3) rendering prognostic information of clinical relevance, through the assessment of molecular predictors of outcome; and (4) assisting in the selection of individualized treatment regimens, thereby gaining specificity and avoiding unnecessary drug toxicity. Companion diagnostic tools are designed to guide targeted clinical trials and personalized oncological treatments. The clinical services rendered also include the analysis of minimal residual disease and evidence of early relapse usually driven by new resistant clones of cancer cells. Protocols based on molecular markers have been proven to increase the chances for cure by opting for the right management approach and improve the quality of life of patients with cancer.
Molecular pathology also serves as a bridge between clinical and basic biomedical sciences. The translational aspects of research in molecular pathology are bidirectional, involving both the active transfer of relevant observations that are the result of the analysis of primary tumors and clinicopathological correlations to basic laboratory studies and the effective transfer of laboratory findings into clinical analyses and protocols. Molecular pathology thus facilitates the transfer of biological discoveries into diagnostic, prognostic, and therapeutic applications.
The use of molecular techniques has led to remarkable progress in our understanding of cell growth, differentiation, maintenance of genomic integrity, and programmed cell death, these being key pathways in tumor development and progression.3–5 Certain biological markers, such as alterations of TP53 tumor suppressor gene, correlate with tumor behavior when detected in specific tumor types.8 Similarly, prospective clinical analyses utilizing well-characterized cohorts of patients and properly selected normal and tumor paired samples are needed to better delineate the role of mutations occurring in these genes, as they impact on patients affected with cancer. Discrepancies of reported results aimed at the identification of gene mutations and altered patterns of gene expression in primary tumors can be explained by the use of different probes and methods. Critical caveats include the presence of viable tumor, the extent of necrosis, normal-to-tumor cell ratio, and the stage and grade of the lesion being studied. Distinct methods are often available to detect a given marker, differing in specificity, sensitivity, speed, and cost, as well as appropriateness for particular clinical situations. The application of the full spectrum of molecular methods to evaluate tissue specimens and the potential availability of therapeutic agents aimed at specific molecular targets have led to changes in the established patterns of tissue procurement, processing, and tissue banking. Protocols should be implemented and constantly updated to guarantee that samples are handled in a way that will allow optimal application of molecular testing. In daily practice, this means that part of a specimen should be set aside prospectively for advanced diagnostics whenever possible. Adding predictive assays to our diagnostic and prognostic tools will enhance our ability to design effective treatment regimens.
Bladder cancer as a model of molecular pathology classification
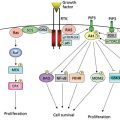
Bladder cancer is the fifth most commonly diagnosed noncutaneous solid malignancy and the second most frequently diagnosed genitourinary tumor after prostate cancer.9–11 Prevalence of bladder cancer is 6 times higher in developed compared to developing countries. As it is characterized by frequent recurrences, bladder cancer represents one of the most costly malignancies to health care systems owing to the requirement for intensive surveillance with cystoscopies and urinary cytologies, as well as frequent tumor resections under anesthesia.9–11 Clinically, most patients with bladder cancer present with dysuria and hematuria.12 After cystoscopy and biopsy evaluation, two distinct superficial (noninvasive) type lesions have been described: low-grade neoplasms, which are always papillary (Ta low-grade urothelial carcinomas), and high-grade carcinoma in situ (Tis or CIS) lesions. CIS is a flat high-grade tumor that progresses to invasive bladder cancer in 60–80% of cases within 5 years, representing the cause of death in 40% of these patients.13–15 These two clinicopathological entities define a novel model for tumor initiation and progression of bladder cancer based on molecular genetic studies.16–22 Two distinct genetic pathways characterize the evolution of early-stage bladder neoplasms, as well as later bladder cancer progression.23, 24 Superficial papillary bladder tumors are characterized by gain-of-function mutations affecting oncogenes H-RAS, FGFR3, and/or PI3K, and deletions of the long arm of chromosome 9 (9q). CIS lesions are characterized by loss-of-function mutations affecting tumor suppressor genes p53, RB, and/or PTEN.25–40 These mutations are the main genetic precursors of invasive bladder cancer. On the basis of these data, a model for bladder tumor progression has been proposed in which two separate genetic pathways characterize the evolution of early bladder neoplasms (Figure 4).

Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree