Abstract
Molecular genomic testing provides clinicians with prognostic and predictive information that can help individualize treatment, decrease the risk of over- or under-treatment, and, accordingly, improve outcomes. In this chapter, we describe several of the genomic tests that are currently available for clinical use in the management of breast cancer. We also discuss ongoing research related to validating and expanding their utility in different patient populations and explain why it is important for clinicians to incorporate these tools into their practice.
Keywords
breast cancer, genomic assays, Oncotype, MammaPrint, prediction, prognosis
Survival after breast cancer diagnosis has improved dramatically over the past 50 years with the introduction and refinement of multidisciplinary treatment regimens consisting of surgical resection, radiation treatment, and systemic therapy. In particular, anthracyclines (e.g., doxorubicin, epirubicin), taxanes (e.g., docetaxel), and now anti-HER2/neu therapies (e.g., pertuzumab, trastuzumab) have been exceptionally effective not only in treating locally advanced breast cancer, but also in enabling less radical surgery through tumor downstaging, prevention of local and distant recurrence, and slowing disease progression in the metastatic setting. However, these agents, particularly anthracyclines, are associated with significant morbidity including but not limited to cardiomyopathy and peripheral neuropathy. In addition, hormonal therapy with selective estrogen receptor (ER) modulators (SERMs; e.g., tamoxifen, raloxifene) and aromatase inhibitors (AIs; e.g., exemestane, letrozole, and anastrozole) has been shown to achieve compelling survival and recurrence outcomes in patients with hormone-responsive disease with side effects that are often better tolerated (at least in the short term) than those of traditional chemotherapy agents. Accordingly, increasing attention has been devoted to improving clinicians’ collective ability to discern which patients are most and least likely to benefit from receipt of systemic chemotherapy.
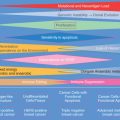
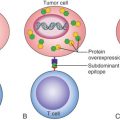
Drawing a distinction between prognostic factors and predictive factors remains relevant with regard to distinguishing models that define risk of recurrence as opposed to those that calculate a response to specific interventions. Prognostic factors are those measurable clinical or biological features of a cancer that provide information about potential patient outcomes before initiation of any therapy. These features undoubtedly reflect inherent tumor biology relating to growth, invasion, and metastases. The major prognostic factors associated with breast cancer include the number of involved lymph nodes, tumor size, histologic grade, and hormone receptor status. For example, the presence of cancer in a locoregional nodal basin is a commonly used and robust prognostic factor, one that is associated with an increased risk of recurrence. In contrast, predictive factors provide information about likelihood of response to a particular therapy ( Fig. 19.1 ). The use of the ER modulators, for example, is important clinically as a predictor of the likelihood of response to hormonal therapy. Frequently, factors may be both prognostic and predictive, blurring the distinction between these two entities. One example of this is the use of HER2/neu, which has significant predictive value for gauging responsiveness to drugs such as trastuzumab, pertuzumab, or lapatinib, but also carries prognostic value in many studies.
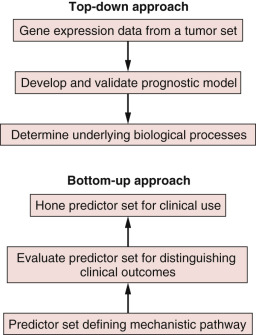
However, our current models of prognostic and predictive factors are quite limited. A large number of tumors recur, despite being found at an early stage, and the ability to better characterize tumors has spurred research for other more robust prognostic and predictive markers. The critical objectives of ongoing research are to develop improved prognostic markers, which are sensitive and specific in their ability to identify individuals who do not require adjuvant treatment, as well as to develop robust predictive markers that will aid in identifying optimal treatment regimens. Molecular genomic assays, which can provide both predictive and prognostic information, have thus emerged as important tools in the pursuit of increasingly personalized treatment for breast cancer.
Genomic assays use tissue samples obtained from a given tumor and facilitate analysis of particular tumor genes that are known to correlate with the natural history of a particular malignancy or the likelihood of that malignancy’s responding to various forms of treatment. These genes may code for a variety of tumor characteristics, from nuclear receptors to tumor suppressors, and their expression can be detected and quantified through various methods from microarrays to immunohistochemistry. There are now several commercially available genomic assays that have been developed to assess the appropriateness of chemotherapy in the adjuvant setting, and they are contributing to increased understanding of the heterogeneity exhibited by breast cancer even among patients who have similar clinicopathologic characteristics. Although they were developed for and are still primarily used in the adjuvant, postsurgical setting, their use in the neoadjuvant setting is also being explored. For this and other reasons, we believe they are important tools with which medical, radiation, and surgical oncologists should be familiar and should feel comfortable ordering for their patients. There are advantages to having the surgeon, as part of a multidisciplinary approach to care, identify the appropriate patient for genomic testing. For example, the time to a decision about the administration of systemic chemotherapy is much shorter when the surgeon orders the test than when it is ordered by the medical oncologist. At our institution, the University of Texas MD Anderson Cancer Center, this window of time was cut approximately in half when the surgeon ordered the test, which has obvious benefits for both the patient and the multidisciplinary team. The sooner genomic testing can be ordered, the more likely it is that patients can consult with their medical oncologist with the results of their genomic testing in hand, so that a firm decision on chemotherapy benefit can be made at the time of their initial consultation. Concerns about the surgeon not being qualified to identify appropriate patients appear unfounded.
Here we review the genomic tests that are currently available and/or in development for clinical use in the management of breast cancer and discuss ongoing research related to validating and expanding their utility in different patient populations ( Table 19.1 ).
| Name | Company | Type | Tissue | Laboratory | Target Population | Outcomes Predicted |
|---|---|---|---|---|---|---|
| Breast Cancer Index | bioTheranostics | 2-gene HI/ five-gene MGI RT-PCR | FFPE | Reference | ER+, LN– | Risk of distant recurrence 5–10 years post-Dx Risk of benefit from 10 years adj endocrine Rx |
| BreastOncPx | LabCorp | 14-gene RT-PCR | FFPE | Reference | ER+, LN– | Metastasis Score: risk of distant recurrence 10 years post-Dx (low, moderate, high) |
| BreastPRS | Signal Genetics | 200-gene microarray | FF, FFPE | Reference | ER+, LN– with intermediate Oncotype Dx RS | Reclassification into high or low risk of distant recurrence 10 years post-Dx |
| EndoPredict | Sividon Diagnostics | 8-gene RT-PCR | FFPE | Local | ER+, HER2– | Risk of distant recurrence 10 years post-Dx (low or high) |
| Genomic Grade Index (GGI) | MapQuant Dx | 97-gene microarray or 4-gene RT-PCR | FF, FFPE | Reference | ER– or ER+, grade II | ER +: Reclassification → low or high risk ER–/ER+: high GGI assoc with ↑ chemosensitivity and ↓ relapse-free survival |
| IHC4 (conventional) | N/A | 4-biomarker IHC | FFPE | Local | ER+ | Risk of distant recurrence 10 years post-Dx |
| IHC4 (NexCourse) | Genoptix | 4-biomarker AQUA | FFPE | Reference | ER+ | Four categories estimating risk of distant recurrence 10 years post-Dx (low, low-mid, mid, and high) |
| MammaPrint | Agendia | 70-gene microarray | FF, FFPE | Reference | ER– or ER+, LN– or LN+ | Risk of distant recurrence 10 years post-Dx (low: <0.4, high: ≥0.4) |
| MammaTyper | BioNTech | 4-gene RT-PCR | FFPE | Local | ER– or ER+, LN– | Improved intrinsic subtyping (especially between luminal A and luminal B) |
| Mammostrat | Clarient | 5-biomarker IHC | FFPE | Local | ER +, LN–, receiving endocrine Rx | Risk of relapse if chemotherapy omitted (low, moderate, high) |
| NPI + | N/A | 4-biomarker IHC; Multivariate model | FFPE | Local | All | Seven biological classes (i.e., refined subtypes) stratified into prognostic groups |
| Oncotype Dx | Genomic Health | 21-gene RT-PCR | FFPE | Reference | ER+, LN–, HER2– | RS: risk of distant recurrence 10 years post-Dx; low: <18, intermediate: 18–30, high: >30 |
| Prosigna (PAM50) | NanoString Technologies | 50-gene microarray | FF, FFPE | Local | ER+, LN–, or LN+, postmenopausal, receiving endocrine Rx | Intrinsic subtyping risk of RS (low, intermediate, high) |
Genomic Assays
Oncotype Dx
First developed in 2004, Oncotype DX (Genomic Health, Redwood, CA) is a 21-gene (16 breast cancer–related genes and 5 reference genes), reverse-transcriptase polymerase chain reaction (RT-PCR) assay that was developed through a multistep process that involved the following:
- 1.
development of an RT-PCR method that could use formalin-fixed, paraffin-embedded (FFPE) samples;
- 2.
selecting 250 candidate genes based on reviews of the published literature and microarray experiments;
- 3.
conducting studies using three independent cohorts of breast cancer patients (n = 447)—participants in the National Surgical Adjuvant Breast and Bowel Project (NSABP)-20 trial (all node-negative [LN–] and ER+), patients with extensive (≥10 involved nodes) axillary disease who received treatment at Rush University Medical Center (Chicago, IL; included both ER+ and ER– patients), and women treated for breast cancer at Providence St. Joseph’s Medical Center (Burbank, CA; included LN+, LN–, ER+, and ER– patients) —to examine the association between the candidate genes and breast cancer recurrence; and
- 4.
using the results of these studies to select the 21 genes for the panel and to develop a genome-based algorithm for predicting recurrence.
The assay was retrospectively validated in 668 tumor samples from women who received adjuvant tamoxifen as part of the NSABP-14 trial, the participants of which all had ER+, HER2/neu nonamplified (HER2–), LN– breast cancer. It was shown to be able to quantify both the likelihood of distant recurrence within 10 years (i.e., is prognostic) and also the likely magnitude of improved distant recurrence–free survival that would occur with receipt of adjuvant hormonal and chemotherapy as opposed to only receiving hormonal therapy (i.e., is predictive). Although not formally approved by the US Food and Drug Administration (FDA), Oncotype DX is currently the only genomic assay recommended in treatment guidelines published by the American Society of Clinical Oncology (ASCO) and the National Comprehensive Cancer Center Network (NCCN) and is also recommended by both the European Society for Medical Oncology and the St. Gallen International Breast Cancer Conference for management of invasive carcinoma. A 12-gene version has also been shown to exhibit predictive and prognostic reliability for ductal carcinoma in situ (DCIS), but the small size of its validation cohort size has limited broader adoption.
Oncotype DX uses FFPE from surgical specimens to categorize patients into one of three tiers based on a calculated recurrence score (RS)—low (<18), intermediate (18–30), and high (≥31–100)—reflecting their likelihood of distant recurrence in 10 years. In the Trial Assigning IndividuaLized Options for Treatment (Rx), also known as TAILORx, women with an RS of less than 11 were found to have a <1% risk of recurrence in 10 years with receipt of endocrine therapy alone, further bolstering support for a paradigm shift away from mandatory chemotherapy within the context of multimodal treatment. Results from the West German Study Group Phase III PlanB Trial provided additional, prospectively generated evidence that patients with an Oncotype Dx RS 11 or less could avoid chemotherapy without compromising outcomes, even if said patients had clinicopathologic characteristics that would otherwise point toward a high risk of recurrence. The RxPONDER Trial (Rx for Positive Node, Endocrine Responsive Breast Cancer) was initiated in 2011 to explore whether ER+, HER2– patients with limited nodal disease (1–3 LNs) and low to intermediate Oncotype Dx scores would experience decreased survival if chemotherapy were omitted from their regimens; another aim of this trial is to determine whether there is an optimal RS cutoff point for these patients, above which chemotherapy should always be recommended. The 21-gene Oncotype Dx assay is mentioned in the NCCN guidelines as a possible consideration to help guide the addition of chemotherapy in patients with limited (1–3) positive nodes because there is ample data from the Southwest Oncology Group (SWOG) 8814, the NSABP B-28, and the other studies just mentioned to suggest that it provides predictive utility of chemotherapy benefit in patients with limited nodal involvement. The eagerly awaited results from the RxPONDER trial will clarify the role for genomic testing with Oncotype Dx in node-positive patients.
In the meantime, Oncotype Dx continues to be the most commonly used genomic test for breast cancer in the United States. Even though the test can be performed on core biopsy specimens, at our institution, we prefer to order the test on the final pathology specimen rather than the core biopsy sample to minimize the chances we will unexpectedly identify multiple positive nodes at operation, thus minimizing the chances of a “wasted” or inappropriate test. Also, Oncotype DX issues separate reports for node-negative and node-positive (N1–3) patients.
MammaPrint
MammaPrint, which was first described in 2006, is a 70-gene DNA assay developed by Agendia (Irvine, CA), a commercial spinoff of the Netherlands Cancer Institute (NKI) and Antoni van Leeuwenhoek Hospital in Amsterdam. It consists of a customized microarray slide that assesses in triplicate the messenger RNA (mRNA) expression of 70 genes initially identified in 78 tumors from a cohort of T1–2, LN– breast cancer patients all under the age of 55 years at diagnosis and treated at NKI; 50% of these patients were ER+. The assay can use either fresh-frozen tumor samples or FFPE. The MammaPrint Index (i.e., score) ranges from –1 to + 1; tumors with a MammaPrint Index less than 0.4 are classified as having a low risk of distant metastasis in 10 years, and those tumors with scores of 0.4 or higher are at high risk for developing distant metastases in 10 years.
Initially, MammaPrint was internally validated using only 19 tumors, and its development was criticized for the small size of both the reference and test cohorts. However, its prognostic value was subsequently validated through a retrospective series using 61 node-negative patients from the initial reference group as well as 144 new node-positive patients and 90 new node-negative patients and was found to better predict 5-year overall survival and the likelihood of developing distant metastases at 5 years than the clinicopathologic risk criteria for recurrence found within the then-current guidelines of both St. Gallen and the National Institutes of Health; 77% of the 295 patients in this study were ER+. Retrospective validation was again performed as part of the international multicenter trial by the TRANSBIG consortium, which included 302 T1–2, LN– patients diagnosed before 1999 who were less than 61 years old at diagnosis and whose treatment was limited to locoregional therapy (i.e., did not receive systemic therapy); 70% of these patients were ER+. The trial was conducted to determine which of three candidate microarrays—MammaPrint, the 76-gene Rotterdam/Veridex signature, or the Genomic Grading Index—should be selected for prospective validation in what would eventually become the MINDACT trial. No significant difference was found between the three methods, and the TRANSBIG consortium ultimately decided to use MammaPrint for prospective validation.
The primary objective of the MINDACT trial, which was launched in 2007, was to determine whether patients with ER+ or ER– disease and a low MammaPrint score but who were deemed high risk for recurrence according to traditional clinicopathologic characteristics (as determined through use of Adjuvant Online!) could safely be spared chemotherapy. It was estimated that 10% to 20% of women who would have received adjuvant chemotherapy based on clinicopathologic criteria would be able to forgo systemic therapy without having any adverse effect on their survival. A total of 6693 patients were enrolled in the study between 2007 and 2011 across nine countries; 80% were LN–, 88% were hormone-receptor (HR)-positive (HR+), and 10% were HER2/neu-amplified (HER2+). Patients with concordant high-risk (n = 1806) and low-risk (n = 2745) assessments underwent chemotherapy or did not receive chemotherapy, respectively. Of the remaining patients with discordant evaluations, 592 were deemed low risk by Adjuvant Online! and high risk by MammaPrint, whereas 1550 were deemed high risk by Adjuvant Online! and low risk by MammaPrint. Among the latter cohort of 1550 patients, 748 were randomized to receive no chemotherapy, and of these 748 patients, 644 were confirmed to have no change in risk postenrollment and therefore received no chemotherapy. The primary analysis of these 644 patients in the MINDACT study was presented at the 2016 American Association for Cancer Research meeting, and the full results were published in 2016. The authors reported that after a median follow-up of 5 years, distant metastasis–free survival was greater than 94% in the patients with discordant evaluations regardless of the treatment arm to which they were randomized. Also, 48% of the patients in the group deemed high risk using Adjuvant Online! and low-risk according to MammaPrint had involved lymph nodes. Thus MammaPrint might show promise as a reliable prognosticator for breast cancer patients, regardless of ER or LN status. The study authors found no added value for MammaPrint in patients who were identified as clinically low risk but had a high MammaPrint result. Notably, the MINDACT study was not powered to predict differential responses to chemotherapy, and the results of the trial should be understood in that context.
Mammostrat
First launched in 2010, Mammostrat (Clarient Diagnostic Services, Aliso Viejo, CA) is a five-biomarker, immunohistochemistry (IHC) assay that measures levels of SLC7A5, HTF9C, p53, NDRG1, and CEACAM5 in FFPE tumor samples to stratify patients receiving endocrine therapy for HR+ tumors into three groups, low, moderate, and high, that reflect risk of relapse if chemotherapy is omitted from adjuvant treatment. It has been retrospectively validated in multiple cohorts of patients with ER+, ER–, LN+, and LN– breast cancer; however, its application in the United States remains limited, and it is not approved by the FDA.
Prosigna Breast Cancer Prognostic Gene Signature Assay
Prosigna (formerly called the PAM50 test; NanoString Technologies, Seattle, WA) is based on a 50-gene RT-PCR microarray (PAM50 test) that uses its proprietary nCounter digital technology to process postoperative FFPE samples of invasive carcinoma and assign tumors to one of four intrinsic subtypes: Luminal A, Luminal B, HER2+, and Basal-like. In addition, the Prosigna gene signature also generates an individualized risk of RS (high, intermediate, or low) representing an estimate of the likelihood of developing recurrent disease through an algorithm that takes into account intrinsic subtype, correlation between molecular subtype and a subset of proliferative genes, and tumor size on final pathology. It has been retrospectively validated in postmenopausal women receiving adjuvant endocrine therapy for both LN+ and LN– breast cancer and was cleared by the FDA for marketing as a prognostic tool in 2013.
Breast Cancer Index
The Breast Cancer Index (bioTheranostics, San Diego, CA) represents a combination of two diagnostic tests, the two-gene, HoxB13/IL17BR ratio index and the Molecular Grade Index, a real-time RT-PCR, five-gene microarray assay. It has been retrospectively validated to predict the likelihood of late (i.e., 5–10 years after treatment) recurrence as well as the likelihood of benefit from a 10-year course of adjuvant endocrine therapy in women with early-stage, LN–, ER+ breast cancer. Specimens can be FFPE or fresh-frozen. It is not currently approved by the FDA for marketing in the United States.
EndoPredict Test
The EndoPredict Test (Sividon Diagnostics, Köln, Germany) combines EndoPredict, an eight-gene, mRNA-based assay that uses RT-PCR on FFPE tumor samples, with patients’ tumor size and nodal status to assign patients with early-stage, ER+, HER2– breast cancer, a score that reflects the likelihood of distant recurrence within 10 years of diagnosis. Patients with a score less than 3.3 are at low risk for recurrence, and those with a score of 3.3 or higher are at high risk for recurrence. The EndoPredict Test is not currently approved by the FDA for marketing in the United States but is approved for use in Europe.
Genomic Grade Index
The Genomic Grade Index (GGI; MapQuant Dx, Ipsogen, France) is a DNA microarray-based assay that uses FFPE tumor samples to measure the expression of 97 genes and assign the tumor a molecular grade. The assay was developed by comparing the gene expression profiles of grade I (i.e., low grade, well-differentiated) and grade III (i.e., high-grade, poorly differentiated) tumors and has also been streamlined into an RT-PCR version that can also use FFPE samples. The test reclassifies grade II (i.e., intermediate grade) ER+ cancers into high- or low-grade categories and thereby confers significantly different prognoses on otherwise similar tumors. High GGI is associated with decreased relapse-free survival in patients who do not go on to receive adjuvant chemotherapy and is also associated with increased sensitivity to neoadjuvant chemotherapy in both ER– and ER+ patients.
IHC4
The IHC4 assay incorporates a semiquantitative assessment of ER, PR, HER2, and ki67 expression using IHC with clinicopathologic factors into a multivariate model for predicting risk of distant metastasis. As originally described, it uses FFPE samples, can theoretically be performed locally, and is a potentially cost-effective method of improving prognostication of early-stage breast cancer with a validated recurrence risk signature. However, its accuracy may be difficult to reproduce in clinical practice given the interobserver variability in IHC assessment, especially with regard to ki67.
The NexCourse IHC4 assay (Genoptix, Carlsbad, CA) purports to minimize this potential variability through use of its internally developed Automated Quantitative Analysis (AQUA) technology for quantification of ER, progesterone receptor, and ki67 expression, although HER2 expression continues to be assessed using IHC or fluorescence in situ hybridization in their assay. In the recently published results of the OPTIMA Prelim trial, there was no significant difference between the conventional IHC4 assay and NexCourse IHC4 with regard to risk assessment for women with ER+ breast cancer. The applicability of this method to clinical practice continues to be a subject of investigation.
Nottingham Prognostic Index/NPI+
The Nottingham Prognostic Index (NPI) is a clinical tool that has been used for more than 30 years to predict prognosis after breast cancer diagnosis. Using a formula that incorporates tumor grade, tumor size, and nodal involvement, patients can be categorized into one of four groups associated with different overall survival estimates, with higher NPIs being associated with worse likelihood of survival at 5 years. More recently, a more granular method of molecular subtyping using 10 vetted biomarkers has expanded the four-tiered intrinsic subtype system, resulting in the identification of seven new breast cancer subtypes: three luminal (Luminal-A, Luminal-N, and Luminal-B), two basal (Basal p53 altered, Basal p53 normal), and two HER2+ (HER2+/ER+, HER2+/ER–). The NPI formula, individualized for each subgroup to only include the most significant clinicopathologic prognostic factors, was then used to further stratify these seven subgroups into prognostic groups, thereby generating a new prognosticator, NPI+ that may prove helpful in clinical decision-making.
MammaTyper
MammaTyper is an in vivo diagnostic test (BioNTech, Mainz, Germany) launched in 2015 that categorizes tumors into intrinsic subtypes through quantitative measurement of ER, PR, HER2, and ki67 using RT-PCR of mRNA from FFPE samples. It was developed to improve discrimination between luminal A and B subtypes. The accuracy of subtype classification with MammaTyper has not yet been compared with PAM50 or IHC, and it is not currently approved for use by the FDA.
BreastPRS
BreastPRS (Signal Genetics, Carlsbad, CA) is a molecular assay that uses an algorithm based on 200 genes sourced from a meta-analysis of publicly available genomic databases to stratify patients into groups at low or high risk for recurrence. It can use RNA extracted from either fresh frozen or FFPE samples and was shown to be able to reclassify patients with intermediate Oncotype Dx RS into low- and high-risk designations. It has not been validated in further studies.
BreastOncPx
The Breast Cancer Prognosis Gene Expression Assay (BreastOncPx, LabCorp, Burlington, NC) is a 14-gene RT-PCR assay that uses FFPE samples to assign patients with ER+, LN– breast cancer a low-, moderate-, or high-risk metastasis score that represents an estimated risk of distant metastases at 10 years after diagnosis. It has not been validated in any further studies.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree