Promyelocytic Leukemia-Retinoic Acid Receptor-α
One of the most elegant examples of the interaction between clinical and molecular advances in the treatment of acute leukemia is APL. The association between the t(15;17)(q24.1;q21.1) translocation and the characteristic morphology of APL (hypergranular blasts with frequent Auer rods or microgranular variant with dumbbell-shaped nuclei) has been known for a long time. The ability to treat APL with retinoic acid (RA) and the understanding of the molecular basis for this treatment is a compelling example of the power of molecular medicine. The initial report from China
37 that all-
trans retinoic acid (ATRA) could induce complete remission (CR) in APL patients actually preceded the discovery that the t(15;17) translocation involved the
RARA gene on chromosome 17.
38,39,40Of four translocations associated with APL, the most common is t(15;17)(q24.1;q21.1), in which the 5′ portion of the fusion protein is encoded by the
PML gene from 15q24.1 and the 3′ portion is encoded by the
RARA gene from 17q21.1. The
RARA gene is a ligand-dependent steroid receptor that mediates the effects of the ligand, RA, on the cell. The breakpoint is invariant in intron 2 of
RARA, yielding the C-terminal portion of the fusion protein, which includes the DNA-binding, ligand-binding, dimerization, and repression domains of RARA. There are three major breakpoints in the
PML gene. The most common generates
PML(L)-RARA, which includes the first six exons of
PML encoding 554 amino acids of PML.
41The wild-type RARA is a nuclear receptor that acts as a transcription factor and binds to retinoic acid response elements (RAREs) in the promoters of many genes, including genes important in myeloid differentiation. RARA binds as a heterodimer with retinoid X receptor protein (RXR) and acts as a transcriptional repressor until ligand (RA) binding occurs, changing the conformation of the protein and resulting in transcriptional activation.
42 Target genes important for myeloid differentiation include colony-stimulating factors (granulocyte colony-stimulating factor [G-CSF]), colony-stimulating factor receptors (G-CSFRs), neutrophil granule proteins (leukocyte alkaline phosphatase, lactoferrin), cell-surface adhesion molecules (CD11b, CD18), regulators of the cell cycle, regulators of apoptosis (BCL2), and transcription factors (RARs, STATs, HOX genes) (reviewed in
Ref. 43). Expression of a dominant negative RARA in either a murine hematopoietic cell line or primary murine bone marrow cells, followed by stimulation with granulocyte-macrophage colony-stimulating factor (GM-CSF), results in arrest of granulocytic differentiation at the promyelocyte stage.
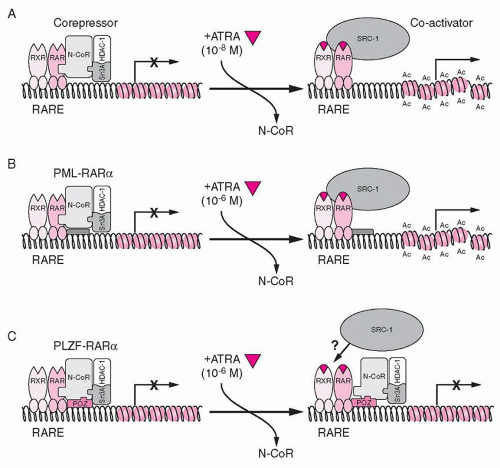
44In the absence of RA, the wild-type RARA, present as a heterodimer with RXR on the RARE, binds to the corepressor proteins SMRT, N-CoR, mSin3, and histone deacetylases (HDACs). Deacetylation of the histones at the target gene promoter results in transcriptional repression. Ligand binding at physiologic concentrations of ATRA causes a conformational change that results in release of corepressors and recruitment of a coactivator complex (SRC-1), which associates with histone acetyltransferases (
Fig. 72.3A).
45 Acetylation of the histones at the target gene promoter is associated with transcriptional activation (reviewed in
Ref. 43).
Wild-type PML protein is normally localized in subnuclear PML oncogenic domains, also called nuclear bodies (NBs), in which other nuclear factors colocalize.
46 PML may act as a tumor suppressor protein and is involved in growth suppression as well as in induction of apoptosis (reviewed in
Ref. 43). Although it does not bind DNA directly, it influences transcription by interacting with both CREB-binding protein (CBP),
47 a transcriptional activator, and HDACs, transcriptional repressors, possibly within the NBs. The protein encoded by the
PML-RARA fusion transcript resulting from the t(15;17) is delocalized from the NBs to a microspeckled nuclear pattern.
48In APL, the PML-RARA protein binds to RAREs with similar affinity to the RARA protein and is able to heterodimerize with RXR. It acts in a dominant negative manner, competing with wild-type RARA for binding to the RAREs. It binds corepressor proteins in the absence of ligand (via the RARA portion of the protein). However, physiologic levels of ATRA (10
-8 M) are not able to convert PML-RARA into a transcriptional activator; pharmacologic concentrations are required (10
-6 M;
Fig. 72.3B).
45,49 This provides the mechanistic basis for the efficacy of treatment of APL patients with ATRA to include differentiation of the promyelocytes.
Understanding of the mechanism of the response of APL to ATRA was furthered by studies of an alternative translocation, t(11;17)(q23;q21.1), which is rarely seen in patients with APL.
50 Patients with this translocation are resistant to treatment with pharmacologic doses of ATRA. The fusion partner gene on chromosome 11q23 encodes
ZBTB16 (previously known
as promyelocytic zinc finger), a transcriptional repressor. The N-terminal portion of the fusion protein encoded by
ZBTB16 includes the N-terminal POZ/BTB protein interaction domain, transcriptional activation and repression domains, and a variable number of zinc fingers important for protein and DNA interactions (reviewed in Refs. 43,45). ZBTB16 interacts with N-CoR, SMRT, mSin3A, and HDAC1 via the POZ/BTB domain,
51,52 and therefore contributes a second binding site for corepressor proteins. Therefore, although pharmacologic doses of ATRA induce release of corepressors from the RARA portion of the fusion protein, the corepressors binding to ZBTB16 are unaffected (
Fig. 72.3C).
43,53 Significantly, concomitant treatment of cells with HDAC inhibitors such as trichostatin A (TSA) restores ATRA sensitivity, since TSA inhibits the deacetylase activity of the corepressors on the ZBTB16 moiety.
49,52Subsequent studies have demonstrate that PML-RARA recruits the polycomb-repressive complex 2 (PRC2) to the promoters of its gene targets. The PRC2 has a H3K27 methylase activity and can initiate gene repression through trimethylation of H3K27.
54 ZBTB16-RARA additionally recruits polycomb-repressive complex 1 (PRC1); treatment with RA releases PRC2 from both PML-RARA and ZBTB16-RARA, but does not release PRC1 from ZBTB16-RARA.
55
Core-Binding Factor Translocations
The t(8;21) is present in approximately 15% of patients with acute myeloid leukemia,
56,57 and the
RUNX1 (runt-related transcription factor 1, formerly called
AML1) gene, cloned from the t(8;21) (q22.3;q22) breakpoint,
58,59 is mutated in another 3% of AML. The activity of the murine counterpart of RUNX1 was first described as part of the core-binding factor (CBF), which binds to a core enhancer sequence of the Moloney murine leukemia virus long terminal repeat.
60 Another component of CBF, the non-DNA-binding CBF
β was found to be associated with inversion 16 in AML.
61 Finally, the fusion partner of
RUNX1 in t(8;21), named
RUNX1T1, or
ETO (eight-twenty-one), also encodes a transcriptional regulator.
62 A gene related to
RUNX1T1, CBFA2T3 (or
MTG16), is involved in yet another translocation involving
RUNX1, t(16;21).
63 The structures of the fusion proteins resulting from these CBF translocations are shown in
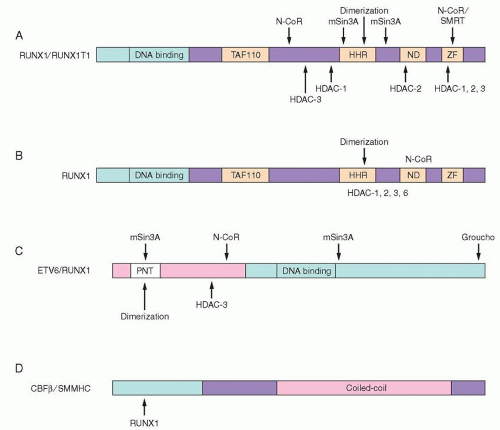
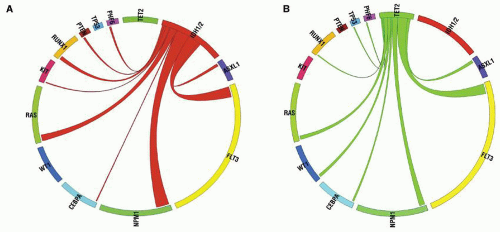
Figure 72.4.
RUNX1 is located at chromosome 21q22.3 and is encoded by 12 exons over 260 kb of DNA. The N-terminal portion of the protein contains the runt homology domain (RHD), which is homologous to the
Drosophila runt protein
64 and is responsible for the official HUGO name,
RUNX1. This is the DNA-binding domain and it is mutated in familial platelet disorder (FPD) and in AML associated with
RUNX1 mutations.
65,66 CBFB interacts via this domain and changes the conformation of RUNX1 to increase DNA-binding affinity.
67 C-terminal to the RHD are potential MAP kinase phosphorylation sites, followed by three weak activation domains, a nuclear matrix target signal, a dimerization domain, and sequences that are recognized by corepressor proteins (reviewed in
Ref. 68).
The CBFs are essential for hematopoietic development. Gene deletion of either
Runx169 or
Cbfb70 in mice results in fetal death at E11.5 to 12.5. These embryos lack all fetal hematopoiesis. Further transgenic experiments have demonstrated that Runx1 is essential for development of HSCs in the aorta/gonadal/mesodermal (AGM) region, the source of definitive hematopoiesis.
71 The essential role of RUNX1 in hematopoietic development appears to be through its function as a transcriptional activator. It regulates lymphoid genes such as B-cell tyrosine kinase, T-cell receptor
α and
β,
72 cytokines (interleukin-3 [IL3],
73 GM-CSF
74), and granulocyte proteins (myeloperoxidase and neutrophil elastase),
75 to name a few. In addition, RUNX1 acts as a transcriptional repressor of genes such as p21Waf1/Cip1 via interactions with the mSin3a corepressor
76 and with SUV39H1, a histone methyltransferase.
77The
ETO gene, now called
RUNX1T1, was cloned from the t(8;21) fusion
58 and is the mammalian homolog of the
Drosophila nervy gene.
78 The four homology domains shared with the Drosophila protein include a region of similarity to TAF110, a hydrophobic heptad repeat (HHR), an ND domain of undetermined function, and two zinc finger motifs that may be a protein-protein interaction domain (
Fig. 72.4A).
68 RUNX1T1 does not appear to bind DNA specifically on its own. However, it may act as a corepressor protein.
79 It associates with N-CoR and mSin3A, and directly binds to the Class I HDACs, HDAC-1, HDAC-2, and HDAC-3 (
Fig. 72.4A).
80In the t(8;21) translocation, the
RUNX1 gene is fused to the
RUNX1T1 gene on chromosome 8. The breakpoint in the
RUNX1 locus is between exons 5 and 6,
81 yielding a fusion protein with the N-terminal 177aa of RUNX1.
58 In this fusion protein, the DNA-binding domain is present, but the C-terminal activation domains, corepressor interaction sites, and nuclear localization signals (NLSs) of the wild type are not present (
Fig. 72.4A).
68 The breakpoint in the
RUNX1T1 gene occurs in the introns between the first two alternative exons of
RUNX1T1, resulting in the inclusion of almost all of the coding region for RUNX1T1 in the fusion transcript.
58The RUNX1-RUNX1T1 protein specifically binds to the same DNA-binding site as RUNX1 and can heterodimerize with CBFB.
82 Therefore, the RUNX1-RUNX1T1 protein can act as a dominant negative inhibitor of wild-type RUNX1. However, cotransfection experiments demonstrated that RUNX1-RUNX1T1 can also function as an active transcriptional repressor, not only inhibiting activation of a reporter gene containing the GM-CSF receptor promoter by cotransfected RUNX1, but also reducing the expression of the reporter gene below baseline.
83 The ability of RUNX1-RUNX1T1 to act as a transcriptional repressor depends on its association with HDACs (via RUNX1T1;
Fig. 72.4A), since the HDAC inhibitor TSA can abrogate effects of RUNX1-RUNX1T1 on the cell cycle.
80 In addition, examination of the M-CSFR gene in Kasumi-1 cells (high expressors of RUNX1-RUNX1T1) reveals an increase in histone H3Lys9 methylation.
84 Targets of RUNX1-RUNX1T1 repression are presumed to include genes important for granulocyte differentiation. In addition, RUNX1-RUNX1T1 represses the tumor suppressor genes
P14ARF and
NF1.85,86 P14ARF stabilizes TP53 by antagonizing MDM2, an inhibitor of TP53.
87 Therefore, repression of
P14ARF reduces the checkpoint control path of TP53, and may be a key event in t(8;21) leukemogenesis. The promoter of
P14ARF has eight RUNX1 DNA-binding sites, and wild-type RUNX1 can activate
P14ARF. However, transfection of
RUNX1-RUNX1T1 into cells that have only low levels of RUNX1 and high endogenous levels of P14ARF results in repression of
P14ARF. Samples of bone marrow from patients with t(8;21) leukemia have low levels of
P14ARF transcript by quantitative real-time polymerase chain reaction analysis.
85 Surprisingly, expression of RUNX1-RUNX1T1 in myeloid progenitor cells inhibits cell cycle progression. However, this may contribute to leukemogenesis by allowing time for accumulation of mutations in a cell immune from TP53-induced apoptosis due to inactivation of P14ARF.
85Finally, inversion 16, present in about 8% of AML cases, involves the CBF complex member CBFB and is associated with a morphologically distinct subset of AML, previously considered M4Eo in the French-American-British Cooperative Group Classification. This disease is a myelomonocytic leukemia with abnormal eosinophils that have dark purple as well as orange granules.
35 This cytogenetic abnormality in which the
CBFB gene is fused to the smooth muscle myosin heavy-chain (SMMHC) gene,
MYH11, results in fusion of the first 165aa of CBFB to the C-terminal coiled-coil region of SMMHC protein (
Fig. 72.4D).
61 A C-terminal region of SMMHC/MYH11 is necessary for the activity of CBFB/MYH11 as a transcriptional corepressor, and this region also associates with mSin3a and HDAC8. Presumably CBFB/MYH11, which cannot bind DNA on its own, interacts with RUNX1 to form a transcriptional repressor complex.
88A number of experiments demonstrate that the CBF translocations are necessary but not sufficient for induction of leukemia. In order to determine whether expression of RUNX1-RUNX1T1 is sufficient to produce leukemia, mice were generated with a conditional
Runx1–
Runx1t1 knock-in allele using the Lox-Cre system. This obviates the embryonic lethality that results when
Runx1–
Runx1t1 is introduced into transgenic mice (recapitulating the phenotype of the
Runx1 null mouse). No leukemia developed in 20
Runx1–
Runx1t1+ mice in 11 months, and no hematologic abnormality was detected except for a slight increase in the number of hematopoietic colony-forming cells. Interestingly, expression of
Runx1–
Runx1t1 did not cause a significant block in differentiation of hematopoietic precursors. When the mice were mutagenized with the DNA alkylating agent, ethylnitrosourea (ENU), 31% of the mice developed granulocytic sarcoma or AML.
89 This supports the hypothesis that several genetic “hits” are necessary for the development of leukemia.
Another study used retroviral transduction of CD34
+ human hematopoietic progenitor cells to investigate the effect of
RUNX1-RUNX1T1 on proliferation and differentiation.
90 In mice reconstituted with RUNX1-RUNX1T1 expressing HSCs there was an expansion of the HSC population and immature myeloid cell populations, although the mice did not develop acute leukemia.
91 Therefore, the expression of RUNX1-RUNX1T1 promotes accumulation of immature cells and prolongs the period of time during which progenitor cells may accumulate additional mutations.
Further support for the hypothesis that genetic mutations besides a mutant
RUNX1 locus are necessary for development of acute leukemia comes from the study of patients with FPD with a propensity to develop AML (FPD/AML). These patients have mutations in one allele of
RUNX1.92 They have defective platelets and progressive pancytopenia, and develop myelodysplasia and a high incidence of AML with age. However, second mutations appear to be necessary before progression to AML occurs. This implies that acquisition of additional mutations is necessary for development of leukemia.
CCAAT/Enhancer Binding Protein-a
CCAAT/enhancer binding protein-
α (CEBPA) is a transcription factor that regulates granulocytic differentiation.
93 Cytogenetically silent mutations of
CEBPA have been identified in about 10% of AML cases.
94 In addition, mutations in other oncogenes in leukemia often lead to
CEBPA downregulation. For example, RUNX1-RUNX1T1 represses the
CEBPA promoter.
95 FLT3-ITD activation of ERK leads to modification of CEBPA which reduces its activity.
96In addition, the
CEBPA promoter is methylated in half of AML cases.
97 The importance of CEBPA in granulocyte differentiation is demonstrated by the lack of mature granulocytes in
Cebpa knockout mice,
98 while its conditional expression triggers granulocyte differentiation in bipotential precursors.
99 CEBPA transactivates the genes for G-CSF and GM-CSF receptors and several granulocyte-specific proteins. The gene produces two proteins using alternative start sites. The larger and predominant 42-kD protein consists of two N-terminal transactivating domains, with a C-terminal bZIP domain consisting of a basic (b) region that mediates DNA sequence binding and a leucine zipper (ZIP) domain that mediates dimerization.
100 The shorter 30-kD protein is transcribed from an alternative internal start site, and retains its bZIP domain but lacks the first transactivation domain. Mutations in
CEBPA are of two types: C-terminal bZIP domain mutations and N-terminal truncating mutations that lead to enhanced production of the 30-kD protein.
101,102,103 The former type inhibits dimerization and DNA binding. The latter type dimerizes with the long form, but inhibits transactivation by the dimer, functioning in a dominant negative manner. In two-thirds of AML with
CEBPA gene mutations, one allele has an N-terminal mutation and the other allele has a C-terminal variant. Several families with familial AML have been documented to have germline
CEBPA N-terminal mutations, and progression to AML has been shown to correlate with a somatic mutation in the C-terminus.
104 CEBPA mutations most often occur in intermediate-risk AML with normal cytogenetics, and these patients have a significantly improved outcome.
94 Interestingly, mutation of
CEBPA at both alleles is associated with a better overall survival than mutation of
CEBPA at a single allele.
105 Approximately one-third of AML with
CEBPA mutations also have
FLT3 mutations, and the
CEBPA mutation confers a more favorable prognosis in this group of AML patients as well.
34
GATA1
GATA1 is a zinc finger transcription factor that regulates erythroid and megakaryocytic differentiation. In the acute megakaryoblastic leukemia (AMkL) that occurs in children with Down syndrome (DS), mutations of
GATA1 have been described in all tested cases.
106,107,108 Familial missense mutations in
GATA1 result in a syndrome of dyserythropoietic anemia and thrombocytopenia, while conditional knockout of
Gata1 in megakaryocyte precursors in mice leads to thrombocytopenia and megakaryoblast proliferation. Approximately 10% of DS patients develop transient myeloproliferative disorder (TMD) in the neonatal period (usually in the first week, almost always within the first 2 months of life), and these patients have mutations in
GATA1.106,109 About one-third of DS patients with TMD later develop AMkL within 5 years, and identical
GATA1 mutations have been identified in the AMkL blasts as were present in the TMD. A large study demonstrated that there is no difference in the
GATA1 mutations present in patients who just developed TMD compared to patients who went on to AMkL.
110 The mutations in
GATA1 result in transcription of a truncated form that lacks its N-terminal transactivation domain, GATA-1s. This shorter form has similar DNA-binding activity but reduced transactivation compared to wild-type, and it therefore can act in a dominant negative manner. By introducing the truncated
GATA1 into
GATA1-deficient fetal liver progenitor cells by retroviral transduction, Muntean and Crispino demonstrated that GATA-1s restored terminal differentiation but that abnormal proliferation occurred.
111 Interestingly, analysis of a knock-in mouse model where
Gata–
1s replaces wild-type
Gata–
1 demonstrates that fetal liver megakaryocytes abnormally proliferate, but that megakaryocyte proliferation is normal in adult bone marrow.
112 This may explain why TMD occurs in the neonatal period in DS patients. Research into the gene(s) on chromosome 21 that cooperate with mutant
GATA1 to produce AMkL in DS children is ongoing; a recent mouse study reported evidence that Dyrk1a (dual-specificity tyrosine-phosphorylation-regulated kinase 1A) can cooperate with mutant Gata1 to promote megakaryoblast expansion.
113AMkL in Down syndrome is sensitive to cytosine arabinoside/anthracycline-based chemotherapy, with event-free survival rates of 80% to 100%.
114 Interestingly, a putative target of GATA1 regulation is cytidine deaminase (
CDA), which inactivates ara-C by deamination to the inactive uridine arabinoside. Presumably failure of GATA-1s to transactivate
CDA increases the efficacy of ara-C treatment.
111