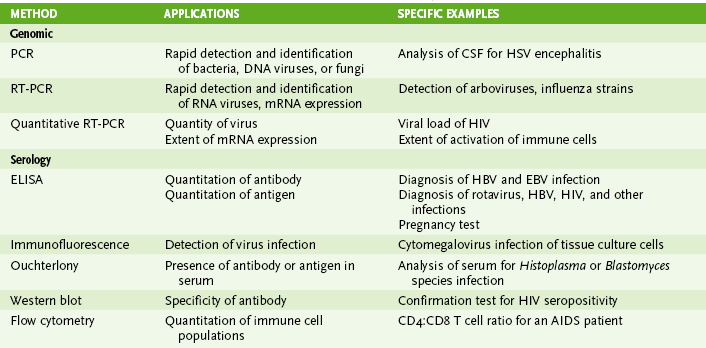
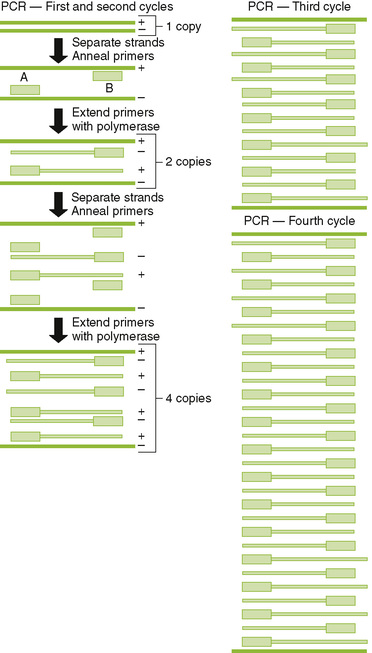
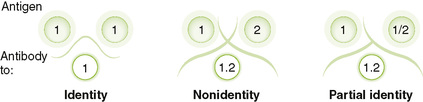
Chapter 5 I Laboratory Assays for Detecting Nucleic Acids (Table 5-1) • Detection of RNA or DNA is rapid and sensitive and can detect microbes that are too virulent or not readily grown in the laboratory. • Methods depend on hybridization of a probe (primer) sequence with a complementary sequence in the sample. • Probes are either radioactive or chemically labeled to allow detection on hybridization with sample. A Southern blotting for DNA and Northern blotting for RNA detect electrophoretically separated genome sequences. B In situ hybridization detects viral DNA or RNA within infected cells. C Polymerase chain reaction (PCR) (DNA), reverse transcriptase (RT)-PCR (RNA), and related technologies permit detection, identification, and amplification of specific nucleic acid sequences. 1. PCR: heat-stabile DNA polymerase amplifies DNA between sequence specific primers (Fig. 5-1) 2. RT-PCR: RT makes a complementary DNA copy of RNA, which is then amplified by PCR using sequence specific primers. • Detects RNA virus genomes, such as HIV • Detects messenger RNA for cytokine genes activated in an immune response 3. Quantitative PCR (qPCR; real-time PCR): rate of production of DNA during PCR reaction determines concentration of DNA in sample. • qPCR after reverse transcription of HIV genome can be used to quantitate the serum concentration of virus (genome load of infection). 4. Other DNA and RNA detection methods include branched-chain DNA assay and antibody capture solution hybridization assay. II Immunologic Assays (seeTable 5-1) • Assays can be used to detect immune responses to infection, or specific antibody can be used to detect soluble and cell-associated antigen. A Antibody-antigen binding (see alsoChapter 3, section IV)
Laboratory Tests for Diagnosis
Laboratory Tests for Diagnosis