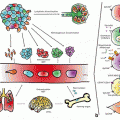
Fig. 1.1
Illustration of the different molecular processes involved in epigenetic mechanisms. (From Qiu 2006; with kind permission of the publisher) .
In mammals, DNA methylation stably alters the gene expression patterns in cells as the basis for a proper and orchestrated realisation of genetic information during development and cell differentiation. Furthermore, it contributes to genome stability, parent-of-origin specific expression of imprinted genes and X-chromosome inactivation in female organisms.
As a result, studying the methylation status of CGIs in a mammalian genome is of major interest for deciphering the regulation mechanisms of gene expression. However, a major challenge is that there is only one genome in one individual, but hundreds of epigenomes as DNA methylation changes during development and is influenced by disease processes and environment (for review: Zhang et al. 2010).
Alterations of specific chromosome regions play, according to recent investigations, a relevant role in the origin and evolution of tumor cells.
In the aberrant tumor karyotype the high amount of intra- an interchromosomal rearrangements can lead to the formation of neocentromeres. This process is regulated by epigenetic mechanisms. The DNA remains unchanged by it has gained an alteration in function.
Furthermore, the centromere of one or more chromosomes might become unstable. This can be caused by hypomethylation of satellite DNA II and III.
Finally, aneuploidisation of tumor cells is a characteristic alteration in cancer. One of the processes which lead to this abnormality is a defect in the checkpoints which regulate the course of the cell cycle.
The Chromosomal Region 11p15 as an Example of an Epigenetically Regulated Segment
The complex spatial and temporal expression of genes regulated by epigenetics can be illustrated by a more detailed description of the genomic region 11p15. The region belongs to the group of chromosomal loci underlying the so-called genomic imprinting , i.e. the parent-of-origin expression of gene-copies. Indeed, inborn imprinting markers are clustered, and approximately 60 imprinted clusters/gene regions have been estimated in the human genome. Among these clusters, the region 11p15 is of central significance as it harbours genes encoding key factors in human development and growth.
The 11p15 region contains a number of imprinted genes whose expression is regulated by two different imprinting centres (ICR1 and ICR2) (for review: Choufani et al. 2013) (Fig. 1.2). The telomeric ICR1 confers a differential chromatin architecture to the two parental alleles leading to reciprocal expression of H19 and IGF2. The two genes are coexpressed in entoderm- and mesoderm-derived tissues during embryonic development and compete for the same enhancers. The paternally expressed IGF2 is a single chain polypeptide and is involved in fetal development and growth. The H19 gene is realised from the maternal allele and encodes a 2.3 kb non-coding mRNA which is strongly expressed during embryogenesis. The physiological function of H19 is unknown but a role as a primary micro RNA in posttranscriptional downregulation of specific mRNAs during vertebrate development has been postulated. The ICR1 contains seven CTCF target sites (CTCF1-CTCF7) in the DMR 2 kb upstream of H19 and shows allele specific methylation. The Zinc-finger binding factor CTCF binds to the maternal unmethylated ICR1 copy and thereby forms a chromatin boundary. This CTCF binding mechanism blocks IGF2 and promotes H19 transcription of the maternal 11p15 copy.
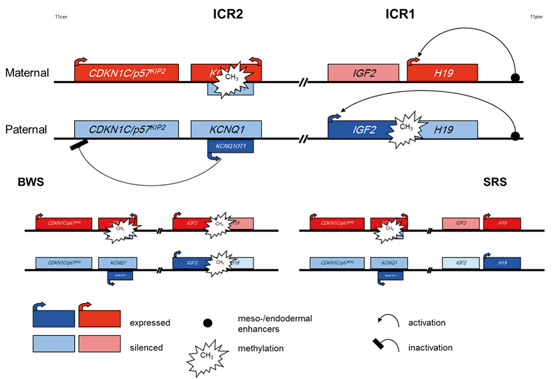

Fig. 1.2
Epigenetic regulation of the two imprinting centre regions (ICR) in 11p15 and illustration of the hyper-/hypomethylation of the ICR1 observed in BWS and SRS as an example for epigenetic regulation in Imprinting Disorders. (for a more detailed description of the complex interactions in these regions. (Choufani et al. 2013)
The centromeric ICR2 regulates the (reciprocal) expression of CDKN1C, KCNQ1 and further genes and is methylated only on the maternal allele. Mutations in the paternally suppressed CDKN1C gene account for up to 40 % of familial Beckwith-Wiedemann syndrome (BWS) cases and 5–10 % of sporadic BWS patients. The gene encodes a cyclin dependent kinase inhibitor (p57KIP2) and is part of the p21CIP2Cdk inhibitor family. Its´ product is expressed in a tissue-specific manner with a 1.5 kb mRNA transcript in the placenta and at lower levels in other tissues. CDKN1C is a mainly maternally expressed growth inhibitor gene that negatively regulates cell proliferation. Overlapping with intron 9, exon 10 and intron 10 of the KCNQ1 locus the untranslated KCNQ1OT1 (LIT1) RNA is encoded. KCNQ1OT1 is expressed by the paternal allele and probably represses realisation of the CDKN1C gene. Loss of methylation of the maternal ICR2 allele correlates with expression of KCNQ1OT1.
Disturbed Epigenetic Regulation
Genomic Imprinting—a Parent-of-Origin Specific Regulation of Gene Expression
As briefly mentioned before, the term genomic imprinting describes an epigenetic marking of specific genes that allows expression from only one of the two paternal alleles (for review: Reik and Walter 2001) . So far, more than 60 human genes are discussed to be imprinted by epigenetic mechanisms but probably there are much more (for review: Horsthemke 2010) . The imprinting marks are inherited from the parental gametes and are then maintained in the somatic cells of an individual. Their programming is subject to a so-called “imprinting” cycle during life which leads to a reprogramming at each generation: During early development, methylation of the mammalian genome runs through dramatic changes and is linked to the rapid differentiation and formation of the various tissues and organs. The imprint marks are erased in the germ-line and re-established according to the sex of the contributing parent for the next generation.
As aforementioned, genes regulated by genomic imprinting mechanisms tend to cluster, thus the imprinting control is often not restricted to a single gene at an imprinted locus but affects the expression of several factors. Due to the numerous factors involved in these complex mechanisms, the balanced regulation of imprinted genes is prone to different disturbances, and indeed several disorders associated with altered genomic imprinting are known, belonging to the group of congenital imprinting disorders (IDs).
Currently eight different imprinting disorders have been reported, among them Angelman and Prader-Willi syndrome. Indeed, the clinical features are different in these diseases, but many clinical as well as molecular overlaps have been reported. Furthermore, similar molecular alterations can be observed in all imprinting disorders:
a.
Aberrant methylation patterns at the differentially methylated regions
b.
Chromosomal duplications or deletions
c.
Point mutations in specific genes (e.g. CDKN1C in Beckwith-Wiedemann and UBE3A in Angelman syndrome)
d.
Uniparental Disomy (UPD).
For the majority of imprinting disorders, patients with uniparental disomy were extremely valuable for the identification of regions harboring imprinted genes. Additionally, it has meanwhile become apparent that UPD can also occur in tumorigenesis.
Disturbed Epigenetic Regulation: 11p15 Alterations as an Example Causing Congenital Imprinting Disorders
For the aforementioned imprinted region 11p15, two syndromes have been identified to be associated with molecular alterations in that region (Fig. 1.2): Beckwith-Wiedemann syndrome and Silver-Russell syndrome (BWS, SRS). Interestingly, the two syndromes show molecularly as well as clinically opposite features: Whereas BWS patients show overgrowth, the main clinical sign of SRS is growth retardation. On the molecular level, different types of mutations and epimutations affecting either the ICR1 or the ICR2 in 11p15 can be observed: In the ICR1, hypermethylation of the H19 promotor and loss of imprinting of IGF2 are detectable in 2–7 % of BWS patients while loss of methylation in the same region is a frequent finding (> 40 %) in SRS. Nearly 50 % of BWS patients show hypomethylation in the ICR2. In 1–2 % of both syndromes, either paternal (BWS) or maternal duplications (SRS) are detectable. Paternal uniparental disomy of 11p15 accounts for 20 % of BWS patients, in SRS only single carriers of maternal uniparental disomy have been reported. In BWS, a genotype-phenotype correlation exists reflecting the functional consequences of the different molecular alterations, whereas in SRS it is too early to define such a correlation (for review: Engel and Antonarakis 2002).
Epigenetics in Cancer
Aberrant methylation patterns are a well-established cause of cancer (Esteller 2008) : hypermethylation of CGIs localized in tumor suppressor genes result in silencing of these genes, whereas hypomethylation leads to activation of oncogenes. Additionally, the well balanced expression of ncRNAs in the affected region is disturbed. Thus, epigenetic mechanisms control cell fate by maintaining a delicate balance between stability and susceptibility to developmental and environmental stimuli. These characteristics make them highly promising targets for molecular diagnostics and drug discovery: epigenetic biomarkers are highly compatible with clinical diagnostic procedures, and they are increasingly used for informing therapeutic decision-making as well as for suitable and personalized therapies for individual patients (Bock 2009) . More details on epigenetic changes as cause of tumor development are given in the following chapter
Loss of Heterozygosity
The aforementioned mechanism of unipaterenatal disomy (UPD) is one possible cause of Loss of Heterozygosity (LOH), a molecular finding which is a characteristic feature of cancer. LOH in general is defined by the functional loss of an allele of a gene of a cell (e.g. a tumor suppressor gene), in which the other allele has already been silenced. In tumor cells copy-neutral LOH can be biologically equivalent to the second hit in the Knudson hypothesis (Knudson 1971) .
Further Examples of Epigenetic Disturbances
For mutations responsible for the fragile X (FraX) syndrome it has been suggested that they interfere with the process of X-chromosome reactivation in oocytes, thus blocking the transcription of loci at or neighboring the fragile site (Xq27.3) and producing the clinical FraX phenotype. However, this hypothesis could not yet been confirmed.
Furthermore, a contribution of altered methylation patterns to the pathoetiology of chromosomal disturbances by gene silencing of proteins interfering with heterochromatin has been discussed (for review: Busson-Le Coniat et al. 1999) .
1.3.1 Cell Cycle and Its Abnormalities in Cancer Cells
Changes in the course of the cell cycle are a characteristic feature of tumor cells and they cause,—depending on the type of disturbance—a broad spectrum of aberrations.
The course of the cell cycle is even in non-transformed meristematic soma cells not identical in all cells but shows tissue-specific peculiarities. But even one specific cells system as the T-lymphocytes of the peripheral blood show differences when comparing different probands caused by the individual genotype or the age. The somatic cell cycle is subdivided into two main subphases that is mitosis and interphase which are classified by their function. In mitosis these are prophase, metaphase, anaphase and telophase, and in interphase these are G1-, S-, and G2. Between the single phases the so-called “checkpoints” are interspersed, which regulate the progression from one phase to the next one.
Furthermore, the cell cycle is characterized by the duplication of the genome (in the S-phase), by the distribution of the chromosomes into two daughter cells in the telophase, and centriol-cycle which regulates the course of mitosis.
The G1-phase of the cell cycle is composed of one chromatid which is replicated during S-phase. This is the phase which leads to the majority of aberrations of tumor cells by the development and evolution of cells with aneuploid or polyploid karyotype.
After the end of the replication the transition into G2-phase can be blocked in tumor cells. In the normal course of the cell cycle this transition is induced by Cyclin and regulated by specific Cyclin-depending kinases. These proteins and their underlying genes are of general relevance in tumor development as they can function as oncogenes or they can inactivate tumorsuppressor genes.
If thus, the transition from S- to G2-phase is not performed correctly, S-phase is passed through a second or even several times, and the resulting karyotype will be polyploid by endoreduplication (4n to 8n).
The abnormal regulation of gene expression during the transition from S- to G2-phase is caused by the missing phosphorylation of Cyclin E and the following degradation and thus leads to an increased replication of the tumor cell.
Aneuploidisation of tumor cells can also be caused by other mistakes in DNA replication.
For example, the centromeric regions of one or more chromosomes can develop instability by hypomethylation of satellite DNA II and III. The consequence is the multiple passing through of the S-phase by the altered chromosomes.
Aneuploidisation of tumor cells can lead to the development of drug resistance in therapy in cases of specific mutations.
Finally, by replication error partial duplications of chromosome segments can originate. The aberrant chromosomes present as triradials. Starting regions of the doubled replication are among others certain fragile sites of the autosomes.
Another phase of the cell cycle causing an increased development of karyotype aberrations is the anaphase of mitosis. The basis of the origin of the different types of genome mutations are abnormalities of the spindle apparatus. It is either multipolar or shows degenerative alterations. If it is not developed at all, in anaphase the two chromatids of each chromosome separate but they are not distributed to the poles of the cell. In the succession either a tetraploid restitution of the nucleus develops or multiple micronuclei are formed.
The development of the structure of the micronuceli is regulated in interphase and mitosis by the centrosome. Its replication is controlled by the protein p53 and when it is missing this leads to the loss of the centriols or their amplification.
Beside the mutations in the gene T53 alterations in the gene STK15 are of relevance, as this gene regulates the centrosome associated kinase and thus plays a role in the aneuploidisation of tumor cells.
A further season for the development of tumor cells caused by an abnormal course of the cell cycle is the missing development of a new cell membrane after the end of mitosis between the two restituted nuclei. This leads to the development of dikaryotic and subsequently even to polykaryotic cells, of which the nuclei can fuse to a polyploidy genome.
A special type of polyploidisation of tumor cells is caused by endomitosis or amitosis, where the replication of the DNA shows a normal course in interphase but the checkpoint from G2 to mitosis is blocked (Cyclin B/CDK1). The membrane of the nucleus is not dissolved and the genome becomes tetraploid.
This pathologic course can be repeated several times and leads to highly polyploid cells.
A characteristic feature of tumor cells is the high instability of the genome. The aberrations which develop are inherited in different frequencies to their daughter cells, depending on their influence on the vitality and the specific length of the cell cycle.
Each pathologic cell can develop into an aberrant cell clone and leads to a “main line” or “side line” of the composite karyotype of a tumor.
Furthermore tumor cells are characterised by an increased number of secondary chromo-some aberrations such as gaps, breaks and exchanges. They are caused by defects in DNA repair. When developing in G2-phase, they present mainly as chromatid aberrations, in G1 phase as chromosome or isochromatid aberrations.
By secondary fusion of heterologous segments in the reactive breakpoints different types of derivative chromosomes can develop.
The development of tumor cells induced by mutations in oncogenes or tumorsuppressor genes can be caused by an increased or reduced production of gene products which then lead to a dysregulation of the cells
Often the cell cycle is significantly reduced in time and this can lead to dramatic increase of specific pathologic karyotypes.
As a rule it can be observed that the cell cycle of tumor cells is changed by different simul-taneously occurring structural and epigenetic mutations which lead to an aberrant course and explain the heterogeneity and the instability during the evolution of the tumor genome (Miller and Therman 2001).
1.4 Tumor Selection and Investigation Methods
Overview on techniques currently used in routine diagnostics in epigenetic disorders. As a huge number of techniques has been reported we can present only those procedures widely applied by many laboratories
1.4.1 Sample Collection and Transportation of Tumor Tissue
Special care is required when taking biopsies, on their transport and their storing, so that their vitality will not be reduced and the necessary investigations can be successfully performed without any restrictions. During the transport of the samples the right temperature (10–20 °C) and pH value must be guaranteed, a suitable buffer must be added to the medium as well as antibiotic and antimycotic solutions.
Before the first use of a new type of transport container they must be tested to exclude that cytotoxic substances are excreted from the plastic into the medium.
Biopsies of tumor probes always afford a maximum of sterile working.
Mailing of tumor samples is possible but the time period should not exceed 3 days. After their arrival at the investigation center they can additionally be kept for some days in the fridge.
If the sample is large enough part of it can be stored by cryoconservation.
Hematologic neoplasias are analysed from peripheral blood samples and bone marrow biopsies, these methods of investigation are well established (Wegner 1999) and usually chromosome analyses are combined with investigations in interphase nuclei by FISH.
1.4.2 Course and Type of Investigations
Vital Biopsies
Depending on the amount of material of the solid tumor sample one or more investigation methods can be chosen.
If the tumor sample is very small (less than 5 mg) or if the tissue shows already degenerative alterations the direct preparation of the cells has to be preferred.
The tumor cells can be analysed by Interphase-FISH (see1.4.4) choosing DNA-probes relevant for the known changes of the tumor type investigated.
Further preferential techniques for the characterization of the tumor are microarray analyses (see 4.5), MLPA (see 4.6) and the different methods for documenting methylation defects (see 1.4.5, 1.4.6, 1.4.7).
If a higher amount of tumor material is available, chromosome analyses are also applied after direct preparation and long term cultivation of the cells. The best quality of the chromosomes for analysis is received after cell culture. Besides, special FISH and MS-MLPA investigations can be performed.
Cryconservation of Tumor Biopsies
The temperature for conservation is − 80 or − 196 °C in the majority of cases. In the latter case the tissue can be stored for several years.
After defrosting the vitality of the cells usually is only slightly reduced and the desirable methods can be applied.
Paraffin Embedded Material
After standard formalin fixation of the tumor tissue it is dehydrated and then embedded in paraffin. If the tumor is small and surrounded by normal somatic tissue the biopsies are cut, transferred on slides, and the tumor tissue is then prepared under microscopic control. Usually histologic investigations have been performed before and add a detailed characterisation of the tumor tissue. Then paraffin is removed and DNA is extracted.
1.4.3 Cell Preparation and DNA Extracts
In Situ Analyses
Interphases and in vivo occuring mitoses are prepared after short time incubation and adding of colcemid as a block of mitoses.
In special problematic cases it might be useful to determine the spontaneous mitotic activity (mitotic index). The number of mitoses available after this procedure is usually small and they show a reduced structural resolution (about 200–250 bands per genome; Shaffer et al. 2013).
Cell Cultivation In Vitro
The sample should not be older than 3 days when the cultivation is started. The tissue can be incubated after enzymatic digestion or it is cut into small pieces (2 × 2 mm).
The culture condition (type or medium, time of incubation and additives have to be chosen depending on the type of tumor).
Note:
While the number of mitoses after long term cell culture is usually high and their quality (bands per genome) good, it has to be taken into account that often, during cultivation, a selection of single cell lines of the tumor occurs or even that normal somatic cells overgrow the tumor cells.
Documentation of Growth Anomalies
Tumor cells show, compared to not transformed cells, a characteristic change of behaviour. They do not grow in vitro two-dimensional in parallel alignment but irregular, partially overlapping each overlapping each other (criss-cross-growth) and, caused by a missing contact inhibition the cell growth becomes more and more three-dimensional (Fig. 1.3, 1.4).
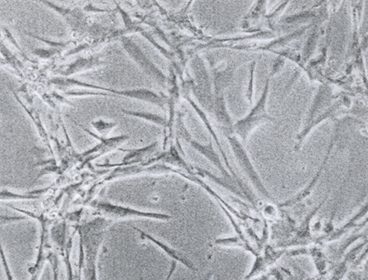
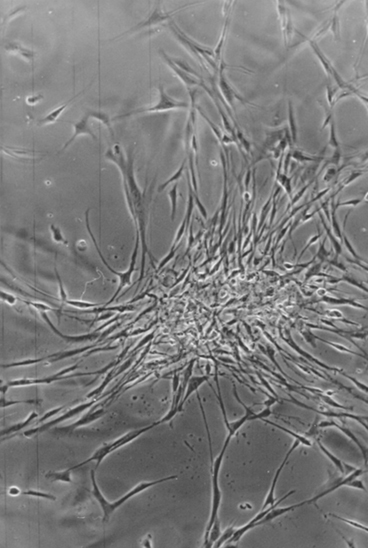

Fig. 1.3
Growth abnormalities in vitro after cultivation of tumor biopsies of a testicular tumor

Fig. 1.4
Criss-cross growth of tumor cells by loss of two dimensional orientation of the cultured cells of a testicular tumor biopsy
The development and extend of these growth anomalies are a relevant parameter characterising tumor cells in vitro.
Characterization of an Abnormal Cell Morphology
In vitro cultivation of tumor cells reveals a high variability of cell size, morphology and nucleus-cytoplasmic ratio. Often cells with long cytoplasmatic appendices are observed. There is an increased number of cytoplasmatic granulae espescially in the surrounding of the nucleus which are an indication for the abnormal metabolism of the cells.
Polykaryotic cells are frequent as well as cells with an abnormal morphology of the nucleus (formation of lobes, perforations, micronuclei). These different anomalies can be observed single or in different combinations and a quantitative documentation has proved to be useful (Fig. 1.5, 1.6).
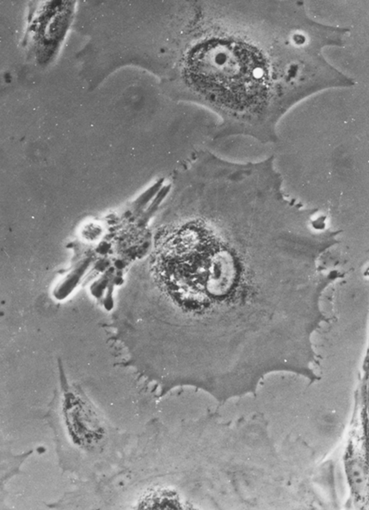
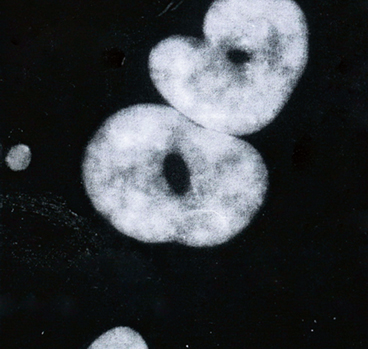

Fig. 1.5
Enlarged polykaryotic cell of kidney tumor with granular inclusions in the cytoplasm

Fig. 1.6
Abnormalities of the nucleus morphology in a kidney tumor with pathologic structure and development of perforations
Preparation of Interphases and Mitoses
The preparation of cells in different stages of the cell cycle is done according to that of normal somatic cells (Wegner 1999) .
1.4.4 Analysis of Cells, Chromosomes and DNA
Structure and Function of the Interphase Nucleus in Normal Somatic and in Tumor Cells
The combination of structure and function of the genome is defined as epigenome. This epigenome consists of different territories localised at defined areas of the interphase nucleus, each of them divided into domains the smallest entities of which are the genes (Haaf and Schmid 1991). The epigenome reveals different types of chromatin condensation and their special arrangements are of high relevance. In this context the celltype investigated as well as the age of the cell also have to be taken into consideration. In euchromatic regions the epigenome influences replication, transcription and repair mechanisms (Cremer 2010) . In the course of the life cycle of a cell the epigenome passes through a number of alterations. As the structure of the nucleus is in tight connection with its function each change in structure has an influence on its function.
Similar to the euchromatin the constitutive heterochromatic regions show characteristic positions in the interphase nucleus. Usually these positions are in the periphery of the nucleus and have the tendency to form heterochromatin clusters. They are defined as chromocenters. As in the euchromatic regions the position and function of heterochromatin can differ in the various somatic cell types.
For example in the early pregnancy extra fetal cells are characterised by heterochromatin which is decondensed, hypomethylated and early replicating whereas in the differentiated placenta the same cell type is highly condensed, hypermethylated and late replicating. Furthermore, the centromeres show in the interphase nucleus a tendency to form clusters and besides a preferential position in the neighbourhood of the inner nucleus membrane. Epigenetic mutations in the centromeric regions influence the correct construction of the different kinetochore elements and thus the separation of the chromatids in the anaphase of the mitosis. Finally, even the telomeres show an increased association in interphases.
The different heterochromatic regions mentioned above have in many cases a relevant function in gene regulation and with reference to guaranteeing the stability of the genome. It could be demonstrated that the stability of the different genome regions is ensured in normal somatic cells by specific factors as the SNF2-family member Fun 30 (Neves-Costa et al. 2009) .
In immortalised cells and in tumor cells the position of the territories in the nucleus is changed (Mai 2013; Wark et al. 2013) . The genome shows a different grouping .
Subsequently, exemplary two types of heterochromatin mutations are given to show the relevance of these changes in the course of tumor evolution.
Changes in the length of the telomeres are known as a characteristic feature in the course of development of normal somatic cells into tumor cells. New techniques enabling 3-D-pictures of the tumor cells in interphase furthermore made it obvious, that telomeres play a role in gene regulations (Mai 2013; Wark et al. 2013). These dynamic regions show changes in their positions in the nucleus of solid tumors, and this change in position was partially reversible. It was the main topic of Mai and co-workers, to find characteristic changes of telomere position in the nucleus, which make it possible to delineate the development of normal somatic cells into precanceroses and finally to tumor cells.
Structural biomarkers can be investigated in vital tumor biopsies, in fixed tissue and in tissue sections.
The centromeres of the chromosomes are characterised by the histon H3 in the variant CenH3. In this chromosome region methylation is the condition for a correct synthesis of the kinetochores. If this reaction does not take place the consequence is a malsegregation of chromosomes (aneuploidisation) which is characteristic for a high number of tumors.
Chromosome Analyses
Chromosome preparations are usually studied after GTG banding.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree