INTRODUCTION
SUMMARY
All T cells express a receptor for antigen that is formed by two polymorphic polypeptides that invariably are associated with a collection of invariant proteins, namely CD3γ, CD3δ, CD3ε, and CD247.* These invariant proteins are necessary for surface expression and signaling by the T-cell receptor. The two polypeptides that form the T-cell receptor on most T cells are termed α and β. A small subset of T cells has receptors formed by different polypeptides termed γ and δ. The polypeptides of the T-cell receptor have a diversity that is comparable to that estimated for immunoglobulin molecules. However, unlike immunoglobulins, the T-cell receptors recognize small fragments of antigen only if they are presented to them by defined major histocompatibility complex molecules on the plasma membrane of another cell, the antigen-presenting cell. The response of the T cell to antigen depends on the intensity of the signal generated by ligation of the T-cell receptor, and is modified by the simultaneous ligation of other accessory molecules. Interactions at the contact sites between T-cells and antigen-presenting cells are organized in the immunologic synapse. The outcome of T-cell antigen recognition can range from immune activation and T-cell proliferation to specific T-cell tolerance and/or programmed cell death.
Acronyms and Abbreviations
AP-1, activation protein-1; APC, antigen-presenting cell; CTLA-4, cytotoxic T-lymphocyte antigen 4; ERK, extracellular receptor-activated kinase; FOXP3, forkhead box P3; ICAM, intercellular adhesion molecule; IFN-γ, interferon-gamma; IL, interleukin; IPEX syndrome, immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome; ITAM, immunoreceptor tyrosine-based activation motif; ITIM, immunoreceptor tyrosine-based inhibitory motif; iTREG, induced regulatory T cell; JNK, c-Jun N-terminal kinase; LAT, linker of activation of T cells; LFA, lymphocyte function associated; MAP, mitogen-activated protein; MHC, major histocompatibility complex; NFAT, nuclear factor of activated T cell; PKC, protein kinase C; PLC-γ1, phospholipase C-1 gamma; RORγt, retinoic acid-related orphan receptor γ thymus isoform; SAPK, stress-activated protein kinase; SH2 domain, Src homology 2 domain; SH3 domain, Src homology 3 domain; STAT, signal transducer and activator of transcription; TFH cell, follicular helper T cell; TGF-β, transforming growth factor beta; Th17, CD4+ T-cell subset that produces cytokines of the interleukin-17 family; TREG, CD4+CD25+ regulatory T cells; V-like, variable-region-like; VLA, very-late activation; ZAP-70, zeta-associated protein of 70 kDa.
*This chapter was written by Thomas J. Kipps, M.D., Ph.D. in the 8th edition and portions of that chapter have been retained.
T-LYMPHOCYTE ANTIGEN RECEPTORS
The structural basis of T-cell recognition of antigen has been known since the 1990s, when a plethora of studies demonstrated that the proteins of the T-cell antigen receptor are structurally related to immunoglobulin molecules.1 The T-cell receptor is formed by a heterodimer, that is, two disulfide-bond-linked polypeptides that are expressed on the cell surface and are associated with a collection of coreceptor accessory and invariant CD3 proteins. In contrast to immunoglobulins, the T-cell receptor is not secreted and also remains membrane bound throughout the activation process. In the majority of T cells, the T-cell receptor heterodimer consists of an α and a β chain, but a small subset of T cells expresses a γδ heterodimer. Following the rule of allelic exclusion, each individual T cell expresses a single α and a single β chain (or a single γ or δ chain, respectively), and can either be αβ or γδ. Each chain is composed of a variable region, consisting of a hydrophobic leader sequence of 18 to 29 amino acids and an aminoterminal domain of 102 to 119 amino acids, and a constant region with a carboxyl-terminal region segment of 87 to 113 amino acids. The variable region is responsible for the variation in the primary structure among different T-cell receptor polypeptides and represents the antigen-binding site, while the constant region is invariant among chains of the same class. Similar to other surface-membrane receptors, each chain is followed by a small connecting peptide, a transmembrane region of 20 to 24 amino acids, and a small cytoplasmic region of 5 to 12 residues at the carboxyl terminus anchoring the polypeptide in the cell membrane. The T-cell receptor chains fold into tertiary structures that are very similar to that of the light and heavy chains of the immunoglobulin molecule. Overall, the structural similarities between the T-cell receptor and immunoglobulins place the genes encoding these receptor proteins in the so-called immunoglobulin supergene family.
More than 90 percent of mature T cells express an αβ heterodimer, making this the major class of T-cell receptor. Without glycan side chains, each α or β polypeptide has a respective size of only 27 kDa or 32 kDa. However, within minutes after being translated into protein, both chains are glycosylated and assembled into a heterodimer composed of a single acidic α glycoprotein of 39 to 46 kDa linked to a more basic 40- to 44-kDa β-glycoprotein via a disulfide bond between the constant regions of the two chains (Fig. 76–1).
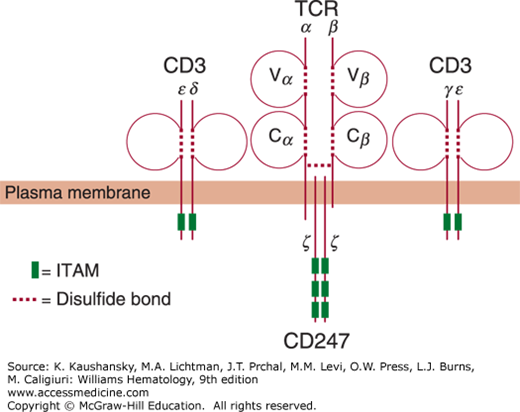
Figure 76–1.
Schematic of the T-cell receptor (TCR) complex. The TCR is composed of the αβ heterodimer, assembled by an α and a β chain, each of which consists of a variable (V) and a constant (C) domain. Both chains form loops and structurally resemble immunoglobulins. The TCR is accompanied by the invariant CD3 molecule, consisting of CD3ε, CD3δ, CD3γ, and CD3. For clarity, CD3ε/CD3δ and CD3γ/CD3ε are depicted separately. An additional component of the CD3 molecule is the ζ chain (CD247) homodimer, which associates with the α and β chain upon stimulation. The dotted lines represent intrachain or interchain disulfide bridges, as indicated in the legend in the lower left-hand corner. The plasma membrane spanned by each of these chains is indicated. The boxes indicate the immunoreceptor tyrosine-based activation motifs (ITAMs) in the cytoplasmic domains of the CD3 polypeptides and the ζ chain.
Less than 10 percent of blood T cells and thymocytes exclusively express a γδ heterodimer. Within the γδ T-cell population, specific γδ T-cell subsets can be defined by their T-cell receptor gene element usage, and their functionally distinct responses to infections with certain organisms such as Listeria monocytogenes.2 Alternatively, γδ T cells can be grouped according to their tissue location. In secondary lymphoid tissues, only 1 to 5 percent of the CD3-positive T cells express γδ receptors. Murine studies demonstrate that many epithelial tissues, such as the epidermis, the intestine, the lung, and the uterus, are enriched for γδ-expressing T cells, indicating that γδ T cells are involved in the surveillance of body barriers.3
The amino acid sequence of the γ chain resembles the T-cell receptor β chain, whereas the amino acid sequence of the δ chain resembles the α chain. Like the homologous αβ heterodimer, the γδ heterodimer is also associated with the CD3 complex and is capable of initiating T-cell activation upon binding of specific ligands. Despite the similarities in chain structure and size, however, the tertiary structure of variable regions of γδ T-cell receptors has a closer resemblance to immunoglobulin variable regions than to the variable regions of αβ T-cell receptors.
Similar to immunoglobulin genes, each chain of the T-cell receptors is encoded by distinct genetic elements that rearrange during development generating a diverse T-cell repertoire (Fig. 76–2).4 Located at band q35 on the long arm of chromosome 7, the β-chain complex has two closely linked genes, each capable of encoding the β-chain constant region. Each constant region gene is associated with a cluster of functional Jβ-gene segments and a single Dβ segment. The functional gene encoding the variable region of the β chain is constructed from the rearrangement of any of approximately 50 variable region gene segments to either one of the two Dβ regions and one of 13 Jβ regions.
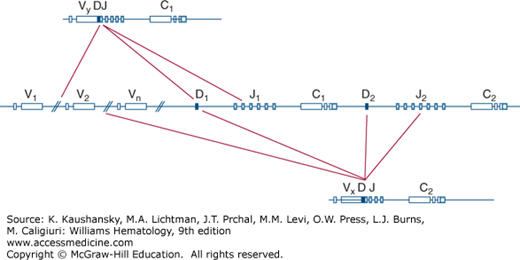
Figure 76–2.
Schematic of possible rearrangements of the T-cell receptor (TCR)-β–chain genes. The TCR-β–chain genes in the germline DNA configuration are depicted in the middle. Possible recombination of either the first constant region (C1, above) or the second constant region (C2, below) with the variable region (V), diversity (D), or joining (J) segments are indicated by the lines.
The α-chain complex is located at band q11.2 on the long arm of chromosome 14 and thus is linked to the immunoglobulin heavy-chain complex. The α-chain gene complex consists of one constant region gene and at least 50 different variable region gene segments. The functional gene encoding the α-chain variable region is derived from the juxtaposition of any one of the variable region gene segments with one of the many Jα segments through rearrangement that generally involves the deletion of the intervening DNA.
The organization of the γ and δ genes is similar to that of the α and β genes, but some significant differences exist. First, the gene complex encoding the δ genes is located entirely within the α-chain gene complex between the Vα and Jα gene segments. Consequently, any rearrangement of the α-chain genes inactivates the genes encoding the δ chain. Second, there are fewer V gene segments in the γ and δ gene complexes than at either the T-cell receptor α or β gene loci. The γ-gene complex on band p15 on the short arm of chromosome 7, for example, has only approximately 12 Vγ gene segments, two virtually identical Jγ segments, and two constant region gene segments. Moreover, there are only approximately four Vδ gene segments, three Dδ gene segments, three Jδ gene segments, and a single constant region gene in the δ gene complex. Consequently, most of the variability in the γ and δ chains is found in the junctional region formed during the process of γδ T-cell receptor gene rearrangement. The amino acids encoded by this region form the center of the T-cell receptor-binding site.
The identification of TCR gene rearrangements is also widely used for diagnostic and therapeutic purposes: In suspected lymphoproliferative disorders, PCR-based clonality testing has been standardized and guidelines and consensus reporting systems have been established.5 Moreover, the molecular analysis for clonal T-cell receptor gene rearrangements can be used to detect minimal residual disease in patients treated for clonal T-cell disorders, such as T-cell acute lymphoblastic leukemia (T-ALL), and guide clinical decision making and provide prognostic information.6,7
Despite their similarities in structure, there are important differences in the way that T-cell receptors and immunoglobulins recognize antigen: immunoglobulins can bind antigens directly, while T-cell receptors generally require that peptide antigens are bound to a molecule of the major histocompatibility complex (MHC) on the surface of another cell.8,9 MHC molecules, also known as histocompatibility antigens, are highly polymorphic glycoproteins, and have immunoglobulin-like structures themselves. There are two basic classes of MHC molecules: Class I MHC molecules generally bind and present intracellular proteins, that is, peptides that are derived from proteins synthesized and degraded in the cytoplasm of the cell. Class II MHC molecules generally bind peptides that are derived from exogenous proteins and degraded in intracellular vesicles. The human histocompatibility antigens HLA-A, HLA-B, and HLA-C are class I molecules, whereas HLA-D antigens DP, DQ, and DR are examples of class II molecules.
MHC class I molecules bind peptides that are usually 8 to 10 amino acids long. Their binding is stabilized by contacts in the free amino and carboxyl termini of the peptide and the peptide-binding groove of all MHC class I molecules. In addition, the peptide-binding groove is closed at both ends. The corresponding binding groove on class II molecules is open at either end, and peptides that bind to MHC class II molecules are generally at least 13 amino acids long. For both class I and class II molecules, the peptide binding groove is located in the central cleft between the two α helices of the MHC molecule. Steric factors, hydrogen bonding, and hydrophobic interactions between the peptide and the MHC molecule stabilize the peptide within this cleft and generate a tertiary structure that is further modified by amino acid residues of both the MHC and the peptide antigen.
MHC molecules are encoded by a family of MHC genes, which are located on chromosome 6 in humans (Chap. 137). MHC genes can be divided into three subgroups, with antigen presentation being encoded by class I (i.e., HLA-A, -B, -C) and class II (i.e., HLA-DP, DQ, DR) genes. Each of these gene loci exists in different alleles, and both maternal and paternal alleles are expressed concomitantly. The particular combination of class I and class II MHC alleles found on an individual chromosome is known as the MHC haplotype. Therefore, the number of different MHC molecules is greatly increased not only by MHC gene polymorphism, but also by codominant expression of MHC gene products. The resulting differences within MHC molecules are primarily found in the amino acids lining the clefts that hold the peptide antigen, allowing the MHC molecules encoded by each allele to bind a distinctive array of different peptides.
Structural studies show that the T-cell receptor can recognize both the MHC-bound peptide and the polymorphic amino acid residues themselves that surround the peptide-binding pocket.10 The specificity of a T-cell receptor (TCR) is defined both by the peptide it recognizes and by the MHC molecule binding it, and TCRs are therefore likely to engage a composite peptide/MHC receptor.11,12 Additional TCR–MHC binding parameters, such as ligands per cell or dissociation time defined by the affinity of antigen, also have an effect on subsequent T-cell activation, but their exact relationship remains largely controversial.13
Some T cells, however, do not recognize MHC-bound peptides. Instead they recognize antigens that are presented by MHC class I-like molecules encoded by genes that map outside the MHC region. One such family of molecules is called CD1. Despite structural similarities with MHC class I molecules, CD1 molecules present larger extracellular peptides in a MHC class II-like fashion, and are also able to present glycolipids to T cells. Glycolipids are components of mycobacterial membranes; in cells infected with mycobacteria, the CD1 molecules are therefore able to bind and present membrane components such as lipoarabinomannan or mycolic acid. T cells that recognize these complexes play an important role in the immune response to Mycobacterium tuberculosis.14,15
As discussed above, structural studies show that γδ TCRs assume a different tertiary structure than αβ TCRs. As a consequence, γδ TCRs are able to recognize a wider variety of ligands, such as bacterial phosphoantigens, nonclassical MHC-I molecules and unprocessed proteins, which distinguishes them from the great majority of αβ T cells.16 Other γδ receptors can recognize determinants presented by CD1 molecules.17 A number of studies have described increased numbers of γδ T cells in a variety of infectious and autoimmune diseases. Therefore, it is speculated that γδ T cells link innate and adaptive immune responses under infectious and inflammatory conditions. In addition, it is hypothesized that γδ T cells might play a role in novel T-cell-based immunotherapy strategies.18,19
TCR diversity is achieved by several mechanisms, some of which are the same as those that generate diversity among immunoglobulin molecules (Chap. 75). The joining of different V (variable), D (diversity), and J (joining) elements to produce a complete V gene, the presence of uncorrected errors made during the recombination of these genetic elements, and the combinatorial diversity afforded by the random pairing of two chains encoded by separated gene complexes all function to enhance the diversity of the T-cell antigen receptor repertoire. An important difference between T cells and B cells, however, is that B cells are capable of undergoing somatic hypermutation (Chap. 75). This process requires expression of activation-induced deaminase (AID) along with other enzymes expressed primarily by B cells within the germinal center of secondary lymphoid tissue during the immune response to antigen (Chaps. 6 and 75).
TCRs do not undergo somatic mutation, probably because of the central role they play in directing host immune defenses: During differentiation, immature αβ T-cell precursors pass through the thymus, where they are educated to distinguish self from non-self by cell-surface proteins of the MHC (Chaps. 6 and 74). Because the ligand for the αβ TCR is processed antigen presented by MHC molecules, close interaction with the MHC might be lost if the variable region of the TCR were allowed to diverge significantly from the inherited germline repertoire. Furthermore, somatic mutation of expressed TCR variable region genes may lead to constitutive T-cell activation to processed self-antigen presented by self-MHC molecules, which could lead to a breakdown in tolerance to self-antigens and autoimmunity.
THE INVARIANT CHAINS OF THE T-CELL RECEPTOR COMPLEX
The CD3 complex of polypeptides and CD247, also known as the zeta chain (ζ chain) of the TCR, are closely associated with and required for the surface expression of the polypeptide heterodimer.20 Unlike the TCR heterodimers, these polypeptides are invariant and are found on all T cells that express αβ or γδ heterodimers. The CD3 polypeptides are designated CD3γ, CD3δ, and CD3ε. The CD3ε chain couples with either the CD3γ or the CD3δ chain to generate heterodimers that each form a tight association with the αβ (or γδ) receptor heterodimer on the T-cell surface (see Fig. 76–1). Each CD3 polypeptide has a negatively charged amino acid in the central portion of the hydrophobic transmembrane region that stabilizes the CD3 complex with the two chains of the TCR. The ζ chain (CD247), on the other hand, forms a disulfide-like homodimer that primarily associates with the two TCR chains and only weakly associates with the CD3 complex. As such, it cannot be coimmunoprecipitated easily with antibodies to the CD3 polypeptides. In addition, the ζ chain lacks a significant extracellular domain (see Fig. 76–1).
The genes encoding CD3γ, CD3δ, or CD3ε chains are clustered on the long arm of chromosome 11 in band q23. CD3γ has a 16-kDa polypeptide backbone that is heavily glycosylated to assume a final molecular mass of 25 to 28 kDa. CD3δ and CD3ε are each 20 kDa in molecular mass. The CD3δ is a glycoprotein consisting of 30 percent carbohydrate. In contrast, CD3ε is not glycosylated. CD3δ and CD3γ are highly homologous at both the protein and nucleic acid sequence level. The nucleic acid sequence of each predicts CD3δ and CD3γ to have typical signal peptides, respective hydrophilic extracellular domains of 79 to 89 amino acids, hydrophobic transmembrane regions of 27 amino acids, and hydrophilic intracellular domains of 44 to 55 amino acids. CD3ε is similar, with a 22-residue signal peptide, an extracellular domain of 104 amino acids, a transmembrane domain, and a comparatively long intracellular domain of 81 amino acids. Each CD3 polypeptide has one immunoglobulin-like domain in its extracellular domain that is defined by an intrachain disulfide bond (see Fig. 76–1), indicating that these polypeptides are members of the immunoglobulin superfamily. However, unlike the αβ or γδ chains of the TCR, there is no variability in the extracellular domains of the CD3 proteins, indicating that these molecules do not contribute to the specificity of antigen recognition.
The ζ chain has no sequence or structural homology to the other three CD3 chains. It is a nonglycosylated protein of 16-kDa molecular mass that is encoded by a gene found on chromosome 1. The ζ chain has only a very short extracellular domain of 6 to 9 amino acids, a transmembrane domain of 21 amino acids, and a long intracellular domain of 113 amino acids.
The cytoplasmic domains of all the CD3 polypeptides and the ζ chain each contain sequences termed immunoreceptor tyrosine-based activation motifs (ITAMs). Each ITAM contains two copies of the sequence tyrosine-X-X-leucine separated by six to eight amino acid residues, in which X represents an unspecified amino acid. The cytoplasmic domains of each CD3 polypeptide contain one ITAM, whereas each ζ chain contains three ITAMs (see Fig. 76–1). These sequences allow the CD3 proteins to associate with cytosolic protein tyrosine kinases following TCR ligation, thus transducing a signal to the interior of the T cell. The cytoplasmic domains of CD3ε and CD3ζ are particularly important in this regard.
Although the major components of the TCR signaling machinery have been identified and mapped, key components continue to be discovered, adding to the complex network of TCR signal transduction. Signal transduction from the TCR heterodimer to intracellular proteins is mediated by the CD3 polypeptides and the ζ chain. Their spatial organization and the mechanisms by which this is regulated are, however, still poorly understood.21 Upon binding to a specific ligand, the TCR αβ (or γδ) heterodimer undergoes steric changes that result in the phosphorylation of the ITAMs of the ζ chain and each of the CD3 polypeptides (see Fig. 76–1). When tyrosine residues in the ITAMs become phosphorylated, they can act as docking sites for adapter proteins or tyrosine kinases, such as the zeta-associated protein of 70 kDa (ZAP-70), which possesses a Src homology 2 (SH2) domain and a Src homology 3 (SH3) domain. Following ligation of the TCR, Src family protein tyrosine kinases (e.g., Lck) are recruited and become activated. This differentially phosphorylates the ITAMs of the accessory molecules in the TCR complex.22 ZAP-70 is recruited to the phosphorylated ITAMs of the ζ chain via its SH2 and SH3 domains, and subsequently becomes activated. Activated ZAP-70 phosphorylates tyrosine residues in the intracytoplasmic segments of surface CD6 and membrane-anchored adapter protein called linker of activation of T cells (LAT), both of which constitute a device for the amplification and diversification of signals that are responsible for most of the responses that result from engagement of the TCR.23
The network of proteins that interact with the LAT signalosome includes SLP-76, phospholipase Cγ1 (PLC-γ1) and GRAP2. Activated PLC-γ1 mediates hydrolysis of phosphatidylinositol-(4,5)-bisphosphate at the cell membrane, which generates the second messengers inositol-(1,4,5)-trisphosphate and polyunsaturated diacyglycerols, leading to a rapid increase in cytosolic free calcium and activation of the θ isoform of protein kinase C (PKC). Cytosolic free calcium binds to calmodulin, an ubiquitous calcium-dependent regulatory protein. The calcium–calmodulin complex activates the cytoplasmic phosphatase calcineurin, which, in turn, catalyzes the removal of an inhibitory phosphate group on the nuclear factor of activated T cells (NFATs) that retains NFAT proteins in the cytoplasm. Removal of the phosphates from NFAT1 and NFAT2 by activated calcineurin allows these transcription factors to translocate into the nucleus, where they enhance transcription of several activation-induced genes, including those encoding interleukin (IL)-2, IL-4, and tumor necrosis factor.24 The importance of this pathway in T-cell activation is underscored by the strong immunosuppressive activity of calcineurin inhibitors such as cyclosporine and FK506, which are commonly administered in the clinic to treat autoimmune diseases and to prevent graft rejection.
In parallel, diacylglycerol in the plasma membrane recruits RasGRP1, a guanine nucleotide-exchange factor (GEF), which acts on Ras to activate extracellular receptor-activated kinase 1 and 2 (ERK1/2). Activated ERK phosphorylates Elk, which, in turn, stimulates transcription of Fos, a component of the activation protein-1 (AP-1) factor that is a necessary component of the transcription-factor complex required for expression of IL-2 and other critical T-cell proteins. LAT-bound SLP-76 also interacts with Nck and with Vav1 to promote reorganization of the actin cytoskeleton, and with FYB to increase the binding of the integrin CD58 (LFA-1) to its ligand intercellular adhesion molecule (ICAM)-1. DYN2, a member of the dynamin superfamily of large guanosine triphosphatases (GTPases), is also recruited by phosphorylated LAT molecules and participates in the generation of filamentous actin. Dedicator of cytokinesis 2 (DOCK2), another GEF, also activates GTPases of the Rac family, which, in turn, activate another mitogen-activated protein (MAP) kinase called p38, and initiate a parallel enzymatic cascade resulting in the activation of yet another MAP kinase called c-Jun N-terminal kinase (JNK), otherwise known as stress-activated protein kinase (SAPK). Activated JNK phosphorylates c-Jun, the second component of the AP-1 transcription factor required for IL-2 transcription. The guanosine triphosphate (GTP)-bound form of Rac also induces cytoskeletal reorganization, thereby facilitating the clustering of the TCR complex, accessory molecules, and other accessory proteins at the site of contact between the T cell and the antigen-presenting cell (APC).
CD4 AND CD8
CD4 and CD8 are glycoproteins that share structural features of other immunoglobulin superfamily receptor molecules. CD8 has two isoforms with different expression patterns and presumably different functions, and is expressed as a CD8α/CD8β heterodimer or as a CD8α/CD8α homodimer.25 These chains are encoded by genes that are linked closely to the immunoglobulin κ light-chain locus at band p12 on the short arm of chromosome 2. The protein sequence of the aminoterminal domains of each CD8 chain shares greater than 28 percent homology with κ light-chain variable regions. Therefore, these domains are called the variable-region-like (V-like) domains. Following this V-like domain, the CD8 molecule has a short region rich in prolines, threonines, and serines that resembles the immunoglobulin hinge region. This region also contains sites for O-linked glycosylation. A hydrophobic transmembrane region anchors the hinge-like region. The CD8 molecule has a 25-amino-acid cytoplasmic tail consisting of highly basic residues. Two cysteines within the V-like domain form a disulfide bridge that stabilizes the immunoglobulin-like fold. An additional cysteine residue is located each within the V-like domain, the hinge region, the transmembrane segment, and the cytoplasmic domain. These cysteines form intermolecular disulfide bridges between two CD8 molecules, thereby stabilizing the CD8α/CD8β heterodimers or CD8α/CD8α homodimers on the T-cell surface. The cell surface CD8 heterodimer shares structural geometry with the heterodimers formed by the pairing of immunoglobulin light and heavy chains.
CD4, on the other hand, is expressed as a monomer on the surface of a subset of peripheral T cells, mononuclear phagocytes and some blood-derived dendritic cells. It is a 55-kDa monomeric glycoprotein that is encoded by a gene that maps to the short arm of chromosome 12. It consists of 5 external domains, a stretch of hydrophobic transmembrane residues, and a highly basic cytoplasmic tail of 38 residues. Similar to CD8, the aminoterminal domain of CD4 also has extensive homology to immunoglobulin light-chain variable regions. However, following this immunoglobulin-like domain is a domain of 270 amino acids that bears little resemblance to other proteins of the immunoglobulin superfamily.
The cytoplasmic regions of CD4 and CD8 are conserved among vertebrates, suggesting that they are essential for the function of these molecules. The cytoplasmic region of CD4 contains five serines and threonines, one or more of which is phosphorylated by PKC upon activation of T cells by phorbol esters or exposure to antigen. Subsequent to phosphorylation, the CD4 glycoprotein is internalized concomitant with T-cell activation. Similarly, the CD8 protein also possesses a highly charged and conserved cytoplasmic domain that may be involved in transmembrane signal transduction.
In addition to MHC antigen presentation, TCRs generally require activation via CD4 and CD8 as coreceptors. Imaging studies and affinity measurements have demonstrated that CD4 and CD8 molecules associate on the plasma membrane with components of the TCR and contribute to antigen recognition and stabilization of TCR–MHC interactions.26 The adhesion between the CD3/TCR complex and the MHC glycoproteins expressed by an APC or target cell is more than 100-fold enhanced by CD8 or CD4, probably by focusing MHC molecules of the APC or target cell onto the T-cell surface, allowing for specific recognition of “processed” antigen that is cradled within the MHC glycoproteins. However, CD4 and CD8 differ in their expression patterns (see section “Helper and Cytotoxic T Cells and CD4+ T Cell Subsets” below) and MHC-binding specificities: CD8 binds to the nonpolymorphic α3 domain of the HLA class I molecule (HLA-A, -B, or -C), whereas CD4 binds to the nonpolymorphic β2 domain of HLA class II molecules (HLA-DP, -DQ, and -DR).27 Therefore, T cells expressing CD4 or CD8 generally recognize antigens presented by class II or class I MHC glycoproteins, respectively. This selectivity is underscored by studies on knockout mice that lack expression of either of these accessory molecules. Mice lacking CD4 or CD8 fail to develop class II-restricted or class I-restricted T cells, respectively, indicating that these coreceptors play essential roles in the maturation of T cells in the thymus. A similar defect is observed in patients with the bare lymphocyte syndrome who have a congenital immune deficiency caused by genetic defects in their capacity to make MHC class II molecules.28 Although patients have normal numbers of B cells and T cells, they have markedly reduced numbers of CD4+ T cells, thus accounting in part for their profound immune deficiency.
In addition to serving as coreceptors, CD4 or CD8 molecules enhance antigen responsiveness by transducing a signal either directly or in concert with the CD3/TCR complex. This is mediated through their interaction with the SRC family tyrosine kinase Lck.29 Lck is noncovalently associated with the cytoplasmic tails of CD4 and/or CD8. When a T cell recognizes a peptide presented by an appropriate MHC antigen, the interaction of CD4 or CD8 with the MHC molecule brings Lck close to the TCR complex. Lck then phosphorylates the tyrosine residues in the ITAMs of CD3 polypeptides and the ζ chain, thereby initiating the receptor signaling required for T-cell activation.
Finally, CD4 also is a cellular coreceptor for HIV.30 Binding of CD4 along with chemokine receptors such as CCR5 or CXCR4 facilitates entry of the virus into host T cells and stimulates them in an antigen-driven immune response.31 Targeting HIV entry/fusion by specific monoclonal antibodies and/or inhibitors is, therefore, an important therapeutic approach in HIV.32 Additionally, CD4 is also of prognostic relevance, as disease progression correlates with depletion of blood T cells that express CD4 (Chap. 81).
T-CELL SUBSETS
T lymphocytes develop in the marrow from a common lymphoid progenitor that also gives rise to B lymphocytes. While B-lymphocyte precursors remain in the marrow, T-cell precursors migrate to the thymus, where they undergo distinct maturation steps and immunologic education. This is accompanied by characteristic TCR gene and surface expression changes of the CD3 complex, CD4, and CD8. At the early stage, thymocytes are double-negative and express neither CD4 nor CD8. This is a highly heterogeneous population, which includes γδ T cells, αβ
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree