Surveillance for Foodborne Diseases
1 Colorado School of Public Health, Aurora, CO, USA
2 Centers for Disease Control and Prevention, Atlanta, GA, USA
Introduction
Food may become contaminated by over 250 bacterial, viral, and parasitic pathogens. Many of these agents cause diarrhea and vomiting, but there is no single clinical syndrome common to all foodborne diseases. Most of these agents can also be transmitted by nonfoodborne routes, including contact with animals or contaminated water. Therefore, for a given illness, it is often unclear whether the source of infection is foodborne or not. For example, Escherichia coli O157:H7 infections can be acquired by ingesting contaminated food or water or by direct contact with infected animals or persons.
Foodborne diseases are important public health problems worldwide. Diarrheal illness, much of which is foodborne, is a leading cause of mortality in young children. Although diarrheal deaths in developed countries have declined, morbidity remains high. Foodborne diseases can also have an important impact on travel, trade, and development. Prevention of foodborne diseases, therefore, continues to be a challenging and critical public health priority. Surveillance systems for foodborne diseases provide extremely important information for prevention and control.
Foodborne-disease surveillance is typically conducted by local, regional, and national public health authorities who gather reports of diagnosed illnesses. By tracking the number of illnesses over time, surveillance systems can identify demographic, geographic, and temporal trends that can be used by regulators, policy makers, health educators, and others to develop and target interventions aimed at reducing the incidence of disease. Through continued surveillance, the success of these interventions can be evaluated. Surveillance is also crucial for detecting outbreaks of foodborne diseases. Fast and effective outbreak investigations are critical to keeping our food safe. Not only can investigators find and remove unsafe food from the marketplace (e.g., by recalling a contaminated food product) but also investigations provide vital information on what went wrong so that regulatory or other changes can be made to prevent future illnesses.
This chapter describes methods for foodborne-disease surveillance including the advantages and disadvantages of different methods for achieving different public health objectives. The role of surveillance systems in detecting foodborne outbreaks is emphasized.
Objectives of Foodborne-Disease Surveillance
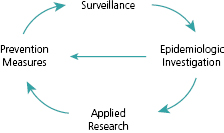
Surveillance is essential to measure, prevent, and control foodborne diseases, as illustrated in Figure 8.1. Information on incidence, trends, and high-risk populations can assist policy makers in prioritizing, monitoring, and evaluating prevention strategies. Surveillance activities can also prompt epidemiological investigations that improve our understanding of disease transmission. The early detection of foodborne outbreaks and their sources can lead to the control of acute public health threats and prevent further spread by, for example, removing contaminated products from the market. Epidemiological investigations can identify gaps in our knowledge about food safety, resulting in identification of new food safety hazards or unsafe food handling practices. These findings often lead to new prevention measures. Thus, the objectives of foodborne-disease surveillance can be summarized as follows: (1) determine the incidence of disease; (2) monitor trends; (3) identify and monitor high-risk populations; (4) detect outbreaks and attribute illnesses to specific sources, including foods, animals, food preparation practices or settings; (5) prioritize interventions; and (6) monitor and evaluate effectiveness of preventive strategies.
Methods for Foodborne-Disease Surveillance
Methods for surveillance can be broadly categorized as (1) passive collection of reports of cases of selected notifiable diseases, (2) monitoring of laboratory-confirmed cases and pathogen subtyping, (3) intensive surveillance in sentinel sites with defined human populations, (4) analysis of hospital discharge records and death registration, (5) review of foodborne-disease complaints systems, and (6) collection of outbreak reports. One method may be more appropriate for meeting certain public health objectives than another. Methods can be used alone or in combination. Most surveillance systems for foodborne diseases conduct surveillance for all diagnosed infections or illnesses and do not routinely attempt to determine the proportion transmitted through food.
Notifiable Diseases
Surveillance of foodborne diseases is frequently part of broader passive surveillance systems that require clinicians and clinical laboratories to report cases of notifiable conditions to the relevant public health authorities [1]. The information collected typically includes the diagnosis, date of diagnosis, and patient demographic information, such as age, sex, and place of residence. Enhanced surveillance to capture additional information on disease severity, symptoms, and food and other exposures (e.g., travel) is often used for severe, low-incidence diseases such as typhoid fever. Timely notification of cases can alert local and national health authorities to potential outbreaks of foodborne diseases; however, dispersed outbreaks may be recognized only when case reports from multiple facilities in a region are collated by a single public health agency. Summaries of reported cases are published at the national (e.g., Morbidity and Mortality Weekly Report in the United States), regional (e.g., Eurosurveillance for the European Union), or global (e.g., The Weekly Epidemiological Record by the World Health Organization) level.
Surveillance for notifiable conditions captures infections only in persons who seek medical attention, receive a diagnosis, and are reported to public health authorities. Systems for surveillance are highly dependent on local clinical and microbiological diagnostic practices, so pathogens for which testing is not routine may be missed (e.g., norovirus, Clostridium perfringens). Reporting delays and undernotification are common. Reports of cases of notifiable diseases are typically collected using passive (or provider-initiated) surveillance. Thus, they rely on the clinician or clinical laboratory to recognize that the disease is reportable and to be motivated to report. This is in contrast to active (or health department–initiated) surveillance systems which contact all providers and institutions that are responsible for reporting cases. Clinicians may be more likely to report if required by law, and reporting is often more complete for serious for rare conditions or for those that may pose an immediate public health threat. Providing clinicians and clinical laboratories with summaries of disease reports and details of any public health action taken raises awareness and may engage further participation in surveillance [2].
Laboratory-Based Surveillance and Subtyping
Surveillance based on laboratory diagnosis generally provides more specific data than surveillance based on syndromes. Laboratory-based surveillance systems typically combine information about the ill person with microbiological information about the pathogen. Standard case definitions and consistent laboratory methods for pathogen identification are essential for effective surveillance of this type. Many foodborne pathogens cause similar symptoms, but laboratory diagnoses identify pathogens. Laboratory-based surveillance can identify both point-source and continued-exposure in regional, national, and international outbreaks. Conducting surveillance for several foodborne pathogens simultaneously can reveal their relative importance.
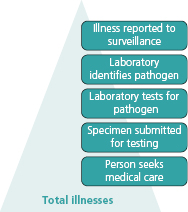
In laboratory-based surveillance only diagnosed and reported illnesses will be captured. As with notifiable disease reporting, the ill person must seek medical care, a specimen must be submitted for laboratory testing, the laboratory must test for and identify the causative agent, and the illness must be reported to public health authorities (Figure 8.2). Many ill persons do not seek medical care, and specimens will not be submitted for laboratory testing for many patients who do seek care. Thus, many cases of the foodborne illnesses go undiagnosed, and most illnesses are not laboratory confirmed [3].
For many years, laboratory diagnosis of foodborne pathogens such as Campylobacter, Escherichia coli O157, and Salmonella, has been based on culture, which is a technique where the sample is placed in a dish with media and watched to see if the bacteria grows. Indeed, most surveillance systems in the United States define a case as a culture-confirmed infection. Recently, however, there has been a shift toward culture-independent diagnostic methods. Although culture-independent tests are usually faster than culture, these tests are generally pathogen-specific so that the physician must make assumptions regarding the causative agent; their performance characteristics (i.e., sensitivity and specificity) are also more variable than those of culture. In addition, because culture-independent tests do not yield an isolate, they may not allow for further characterization of the pathogen.
Further characterization of pathogens by serotyping or molecular subtyping is critical for determining which cases are caused by common pathogens. Antimicrobial susceptibility testing guides treatment and also serves as a form of serotyping.
Serotyping
The introduction of serotyping and molecular subtyping methods has enhanced the detection of low-level, geographically dispersed outbreaks. The first widespread effort at serotyping began in 1962 when clinical laboratories in the United States began sending strains of Salmonella isolated from humans to state public health laboratories for serotyping [4]. Since 1995, the Centers for Disease Control and Prevention (CDC) has routinely used an automated statistical outbreak detection algorithm that compares current reports of each Salmonella serotype with the preceding 5-year mean number of cases for the same geographic area and week of the year to look for unusual clusters of infection [5]. The sensitivity of Salmonella serotyping to detect outbreaks is greatest for rare serotypes, because a small increase is more noticeable against a rare background. The utility of serotyping has led to its widespread adoption in surveillance for food pathogens in many countries around the world [6].
Molecular Subtyping
Today, a new generation of subtyping methods—including pulsed-field gel electrophoresis (PFGE), multiple-locus variable number tandem repeat analysis (MLVA), multilocus sequence typing (MLST), and others—is increasing the specificity of laboratory-based surveillance and its power to detect outbreaks [7]. In the United States, PFGE is used for routine subtyping of E. coli O157, Salmonella, and other enteric pathogens.
Molecular subtyping allows comparison of the molecular “fingerprint” of bacterial strains. In the United States, the CDC coordinates a network called PulseNet that captures data from standardized molecular subtyping by PFGE. By comparing new submissions and past data, public health officials can rapidly identify geographically dispersed clusters of disease that would otherwise not be apparent and evaluate them as possible foodborne-disease outbreaks [8]. The ability to identify geographically dispersed outbreaks has become increasingly important as more foods are mass-produced and widely distributed. Following the launch of PulseNet in 1996, the number of E. coli O157:H7 outbreaks detected nationwide increased dramatically, demonstrating the utility of this approach [9]. Similar networks have been developed in Canada, Europe, the Asia Pacific region, Latin America and the Caribbean region, the Middle Eastern region and, most recently, the African region, facilitating the detection of regional and interregional clusters (www.pulsenetinternational.org).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing is used to monitor the prevalence of antimicrobial resistance in enteric bacteria and can function as a subtyping method for surveillance. Many countries now monitor the prevalence of antimicrobial resistance in Salmonella, Campylobacter
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


