Surveillance for Seasonal and Novel Influenza Viruses
European Centre for Disease Prevention and Control, Stockholm, Sweden
Introduction
Influenza is a vaccine-preventable viral infection that causes systemic and respiratory symptoms. Infected patients present with a wide range of clinical severity. The virus is spread primarily through airborne transmission via droplets and droplet nuclei. Epidemics in humans occur worldwide with marked winter seasonality in temperate regions and a more stable background activity in the tropics [1]. The virus undergoes frequent genetic changes through mutation or reassortment. These may result in minor or major antigenic changes known as drifts or shifts, respectively. Antigenic changes may alter the effectiveness of the influenza vaccine, and therefore vaccines are reformulated annually. Furthermore, major virus antigenic changes may result in influenza pandemics, essentially an epidemic of worldwide proportions. Influenza pandemics are rare events which occur when a novel influenza virus strain emerges and is (1) pathogenic for humans, (2) easily transmissible, and (3) antigenically unique (i.e., pre-existing immunity in the population is low). Because of pandemics’ unpredictability, their potential to cause more severe illness and to affect wider portions of the population, and the difficulties in rapidly producing and deploying an effective vaccine, pandemics are considered major public health threats. Substantial and constant efforts are made by public health authorities to maintain an adequate level of preparedness to effectively respond to influenza pandemics and reduce their impact.
Influenza poses many challenges to surveillance because of its changing nature, its ability to rapidly spread worldwide and affect large portions of the population, the nonspecificity of its clinical symptoms, and the wide range of possible severity and complications. As a consequence, public health surveillance of influenza is often based on collection, integration, and analysis of information from multiple sources and systems. Although each surveillance system has specific objectives, it is only by combining data from multiple systems that it is possible to have an understanding of the burden of disease. Precise estimates of the global burden of influenza are also missing because of the absence of surveillance systems in many low- and middle-income countries. Based on extrapolations of U.S. surveillance data, the World Health Organization (WHO) estimates an annual influenza burden of up to 1 billion infections, 3–5 million cases of severe disease, and 300,000–500,000 deaths [1]. This chapter describes the primary objectives of influenza surveillance and the methods applied to achieve these objectives.
Clinical, Epidemiological, and Virological Characteristics and Implications for Surveillance
Clinical manifestations of influenza include combinations of systemic and respiratory symptoms. Typically patients with influenza report abrupt onset of fever, myalgia, headache, malaise, and anorexia. Respiratory manifestations include dry cough, pharyngeal pain, nasal obstruction, and discharge. Symptoms last for 2–7 days (although cough can persist for weeks) with a wide range of possible clinical severities and complications. Influenza cannot be distinguished from other respiratory infections based on clinical criteria alone [2]. Based on their cause, complications from influenza can be classified into three main categories: (1) directly related to the viral infection (e.g., acute respiratory failure), (2) secondary bacterial infections (e.g., bacterial pneumonia), and (3) exacerbation of pre-existing conditions (e.g., chronic obstructive pulmonary disease, myocardial infarction, stroke). Because clinical symptoms and possible complications of influenza are nonspecific, and because the incidence of disease in the population can be very high during epidemic periods, most influenza infections are not confirmed by laboratory testing. In addition, influenza can cause mild symptoms, and many patients do not consult a physician. This implies that assumptions need to be made when interpreting surveillance data.
There are three types of influenza viruses—A, B, and C— of which only types A and B cause widespread outbreaks in humans. Influenza A viruses are classified into subtypes based on antigenic differences between their two surface glycoproteins, hemagglutinin and neuraminidase. Seventeen hemagglutinin subtypes (H1–H17) and nine neuraminidase subtypes (N1–N9) have been identified. Viruses of all hemagglutinin and neuraminidase subtypes have been recovered from aquatic birds, but only three hemagglutinin subtypes (H1, H2, and H3) and two neuraminidase subtypes (N1 and N2) have established stable lineages in the human population since 1918. Only one subtype of hemagglutinin and one of neuraminidase are recognized for influenza B viruses [3]. The internationally accepted naming convention for influenza viruses contains the following elements: the type (e.g., A, B, C), geographical origin (e.g., Perth, Victoria), strain number (e.g., 361), year of isolation (e.g., 2011), for influenza A the hemagglutinin and neuraminidase antigen description (e.g., H1N1), and for nonhuman origin viruses the host of origin (e.g., swine) [4].
Because the influenza virus undergoes frequent antigenic changes, the vaccine has to be reformulated annually and separately for the two hemispheres. This requires global collaborations to ensure efficient monitoring of strains of viruses circulating in each season and consolidated mechanisms to provide timely recommendations to vaccine producers. Often in the Northern and Southern Hemispheres, annual influenza epidemics occur following certain geographical patterns of spread, thus offering early warning surveillance systems opportunities for informing public health action.
The burden of influenza can be estimated by means of various measures, including numbers of infections, physician office visits, hospitalizations, complications, deaths, and composite burden of disease measures like disability-adjusted life years (DALYs). DALYs estimates account for both premature mortality (years of life lost) and time spent with reduced health (years lived with disability) [5]. Various factors influence the burden of disease including the virulence of the circulating strains, the characteristics of the affected population (level of natural immunity and vaccination coverage, age structure, prevalence of individuals with chronic conditions, proportion living in long-term care facilities) and the setting in which infection occurs (level of access to and quality of healthcare systems) [6]. Use of DALYs to estimate the burden of influenza underestimates the real burden of disease because this measure does not fully account for the whole spectrum of consequences of infection, as is the case for many other infectious diseases [7].
Possible Surveillance Schemes
Virologic Surveillance
Virologic surveillance of influenza has the main objective of monitoring virus strain circulation globally in order:
- to inform the process of vaccine strain selection twice yearly and provide virus seeds for vaccine production;
- to rapidly identify novel viruses that may pose a pandemic threat; and
- to monitor levels of antiviral resistance.
In combination with epidemiological and clinical surveillance, laboratory surveillance contributes to identifying when and where influenza epidemics are occurring and to understanding the relation between virus strain circulation and clinical severity.
Laboratory Diagnosis of Influenza
The most important diagnostic tests for surveillance purposes are reverse transcriptase polymerase chain reaction (RT-PCR) and viral culture. Because of its high sensitivity and specificity, RT-PCR is the preferred test for virus detection and initial characterization by type and subtypes. Results are usually available within less than one day. A subset of positive specimens can be placed in culture for further genetic and antigenic characterization. Although performed less frequently than molecular detection methods, virus isolation through culture provides critical information for surveillance. This includes identification of novel strains that would be classified as unsubtypable by RT-PCR, antigenic characterization evaluating virus relatedness with the recommended vaccine strains, and antiviral resistance testing.
Commercial rapid influenza diagnostic tests are also available and provide results in 10–30 minutes, but they have limited value for surveillance because of their low sensitivity compared with RT-PCR and culture. Preferred respiratory samples for influenza testing include nasopharyngeal or nasal swab or nasal wash or aspirate. Samples should be collected within the first 4–5 days of illness as virus shedding decreases rapidly thereafter, compromising the sensitivity of the tests (i.e., increasing the number of false negative results).
In order to increase representativeness, specimens for virological surveillance should be collected from various sources year round and from patients with a range of clinical severities. Settings where specimens for virological testing can be collected include primary care physicians’ offices, hospitals, nursing homes, occupational settings, and outbreak investigation settings such as schools. It is important to ensure a variety of sources as theoretically some strains can be linked to clinical severity. Because of the potential risk of zoonotic infections and/or emergence of reassorted human/animal viruses, specimens should also be taken from subjects in specific occupational categories, including veterinarians, and in settings where food animals such as poultry and swine are in contact with humans. Even in temperate regions, influenza viruses circulate year round and novel strains can emerge at any time and virological surveillance must be performed year round. At the beginning of each season, the number of specimens collected for laboratory testing must be increased in order to verify vaccine-virus match and to inform the vaccine strain selection process for the following influenza season.
WHO Global Influenza Surveillance Network
At the global level, virologic surveillance is coordinated by WHO through its Global Influenza Surveillance and Response System (GISRS) [8]. The network was established in 1952 and comprises six WHO Collaborating Centres (CCs), four Essential Regulatory Laboratories (ERLs), and 137 National Influenza Centres (NICs) in 107 WHO Member States (Figure 6.1).
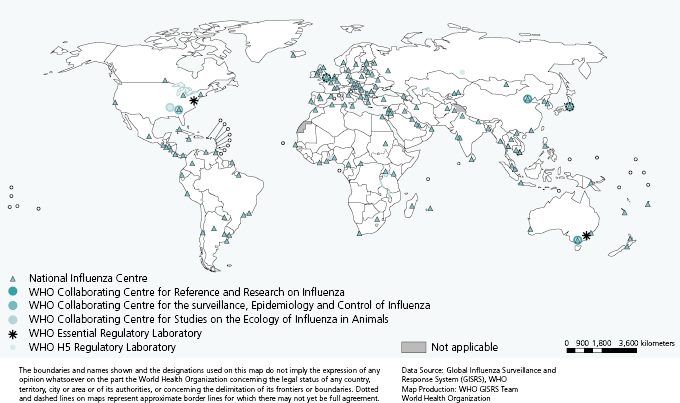
Although NICs’ laboratories represent the first level of the GISRS, their functions, as well as virus detection and characterization capacities, vary depending on the country. In some countries these are the only laboratory facilities in place for influenza virus detection, whereas in others they are part of larger networks of laboratories. According to WHO, NICs should be able to undertake initial identification of virus type and subtype [9]. At the national level one or more NICs receive specimens collected from patients with suspected influenza. Specimens may come directly from primary care physicians, hospitals, and other health institutions or through networks of subnational laboratories. NICs usually receive a combination of specimens, some of which have been collected as part of well-defined surveillance schemes (e.g., from sentinel physicians swabbing patients presenting with an influenza-like illness during the influenza surveillance period).
Vaccine Strain Selection
Influenza strains to be included in seasonal influenza vaccines are selected during WHO consultations, which occur twice a year (in February for Northern Hemisphere and in September in the Southern Hemisphere). The two influenza A strains currently circulating in humans, A(H3N2) and A(H1N1), and an influenza B strain are recommended for vaccine inclusion. Advisors from WHO CCs and ERLs take part, and consultations are observed by multiple experts from WHO CCs, WHO ERLs, WHO H5 Reference Laboratories, NICs, and other institutions. The process relies on the number, quality, timeliness of collection, and geographical representation of analyzed isolates [10–12].
Antiviral Resistance
Only two antiviral drug classes are licensed for chemoprophylaxis and treatment of influenza—the adamantanes (amantadine and rimantadine) and the neuraminidase inhibitors (oseltamivir and zanamivir). Amantadine has been in use since the mid-1960s, whereas the neuraminidase inhibitors (zanamivir and oseltamivir) became available in the late 1990s [13,14]. Antivirals shorten duration of illness, reduce the risk of severe complications, and prevent transmission or reduce disease severity when administered as chemoprophylaxis pre- or postexposure [15]. Antiviral resistant strains arise through selection pressure in individual patients during treatment. Strains selected under the pressure of antiviral treatment may result in treatment failure; however, they usually do not transmit further (because of impaired virus fitness) and have limited public health implications. On the other hand, primarily resistant viruses have emerged in the past decade and in some cases have completely replaced the susceptible strains. This happened for A(H3N2) viruses resistant to adamantanes since 2003 [16,17], seasonal A(H1N1) resistant to oseltamivir since 2007 [18,19], and 2009 pandemic A(H1N1)pdm09 viruses resistant to adamantanes [13]. Antiviral resistance can be tested by genotypic identification of mutations that are known to be associated with drug resistance or analysis of the drug concentration that inhibits neuraminidase activity by 50%.
Surveillance of antiviral resistance has direct implications for the clinical management of patients. Antivirals are most effective if administered within 48 hours from onset of symptom. This short window of opportunity does not allow routine performance of antiviral resistance testing to guide treatment. Clinicians therefore choose among the few available antiviral classes based on updated antiviral resistance prevalence data combined with local data on the relative prevalence of each virus type and subtype [15]. It is therefore important that information on subtype circulation and levels of antiviral resistance is readily available at subnational levels as differences in distribution of subtypes may exist in different parts of a country.
Outpatient Sentinel Surveillance
One of the most important objectives of influenza surveillance is to detect rapidly when and where the influenza season is starting and which virus types and subtypes are circulating and to monitor the geographical progression of seasonal epidemics. This information is used to guide public health interventions at the local, national, and global levels. Examples of interventions may include activating hospital preparedness plans and recommendations for use of antivirals without waiting for the results of laboratory tests. Other measures may be aimed at strengthening ongoing vaccination campaigns and promoting hand-hygiene practices to reduce transmission. Indications of the geographical spread and progression of the annual epidemics allow mobilization of local and national resources and better coordination of efforts to manage demands on the healthcare system, including increased use of intensive care facilities. At an international level, alert mechanisms can be established so that neighboring countries are rapidly informed that the influenza season has started. This information, along with an initial indication of the characteristics of the epidemic in terms of severity, population attack rates, predominant virus types and subtypes, and antigenic and genetic characteristics of viruses and their relatedness to vaccine strains allows countries to prepare accordingly. Surveillance systems based on primary healthcare services are often used to provide this information.
System Based on Sentinel Physicians
In Europe a multinational surveillance network now known as the European Influenza Surveillance Network (EISN) has been operational since 1996; it is currently operated by the European Centre for Disease Prevention and Control (ECDC) for the European Union (EU) and from the WHO Regional Office for Europe for the remaining countries of the European Region of WHO. EISN data is based on sentinel primary care physicians who report numbers of cases of influenza-like illness (ILI) or acute respiratory infections (ARI) observed in their practices during a predefined surveillance period, usually between International Organization for Standardization (ISO) weeks 40 of one year and 20 of the following year. In the EU, ILI is defined as sudden onset of symptoms and at least one of the following four systemic symptoms: fever or feverishness, malaise, headache, and myalgia, and at least one of the three respiratory symptoms (cough, sore throat, shortness of breath) [20]. Weekly aggregate data on ILI or ARI are transmitted with limited epidemiological information, usually age groups and sex. In some cases, presence of risk factors for severe influenza and vaccination status are also collected and reported. Data collected through sentinel networks are analyzed at national and international levels and used for the production of weekly bulletins [21,22].
Sentinel physicians are usually general practitioners and pediatricians who represent 1–5% of primary care physicians, depending on the size of the country. The denominator for calculating weekly incidences is the number of individuals registered with the sentinel physicians or the population under their catchment area. In the absence of reliable denominators, some countries monitor the proportion of weekly consultations for ILI or ARI over the total number of weekly consultations. Aggregated data are transferred to the national level and then to ECDC and WHO using a single online entry form. It is therefore possible to monitor trends of ILI and/or ARI incidence in the community and detect when such incidence increases above certain thresholds. Statistically defined thresholds can be calculated to define epidemic periods using time series analysis techniques such as the moving average method [23].
Integration of Clinical, Epidemiological, and Virological Surveillance
A proportion of sentinel physicians are also asked to collect nasopharyngeal swabs from a subset of patients with ILI. Ideally, patients are selected using a systematic approach to increase representativeness. For example, all ILI cases presenting on a certain day of the week or the first ILI case presenting on predefined days of the week are selected. Swabs are sent to local or national reference laboratories for virus testing, usually using RT-PCR. This allows calculation of the proportion of reported potential cases that are positive for influenza each week. The proportion of samples testing positive for influenza gives an indication of the influenza virus circulation in the community compared to other respiratory infections. Integrated systems allow for the monitoring of the distribution of virus types and subtypes and collection of representative samples and/or isolates for genetic and antigenic characterization. If specimens are collected systematically and if the sampling fraction is known (i.e., the proportion of all observed ILI and/or ARI cases that are positive for influenza), such integrated systems give important information for understanding the burden of disease. Sentinel integrated systems have also been used to monitor the effectiveness of the seasonal vaccines. A proper integration between clinical and virological surveillance is difficult to achieve as it requires that a sufficient number of sentinel physicians are recruited each year, that they are distributed in a geographically representative way, that they are committed to adhere to the surveillance protocol throughout the surveillance period, that a system is in place to ensure rapid transfer of specimens to the reference laboratories, and that there is sufficient laboratory capacity to test specimens collected.
Use of Qualitative Indicators
A number of qualitative indicators are used to describe influenza activity by intensity, trend, geographic spread, and impact. Intensity and trend can be directly derived from the outpatient ILI incidence and can give an indication of the proportion of the population experiencing ILI in a given week and whether such proportion is increasing, decreasing, or stable compared to the previous week (Figure 6.2). Geographic spread describes the extent of virus circulation in a given country or region. It ranges from no activity to widespread activity. Impact refers to the degree of disruption of healthcare services as a result of acute respiratory disease and can range from low (no increased demand) to severe (i.e., shortages of hospital and/or intensive care beds).
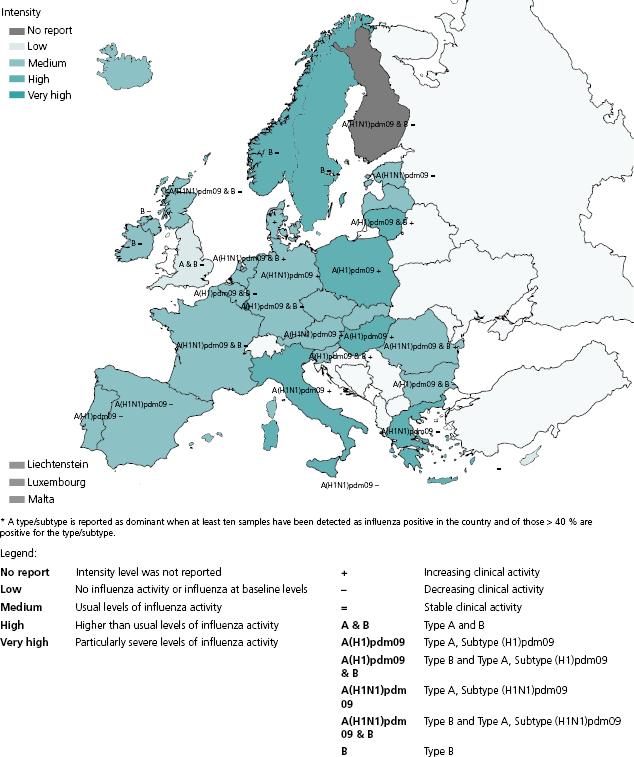
The specific indicators selected for use ultimately represent the judgment of national and or local surveillance authorities, taking into account additional sources of information such as absenteeism rates from schools or workplaces. Reporting of qualitative indicators may be the only option for countries with limited resources. Furthermore, if routine surveillance systems are disrupted because of catastrophic events or increased demand for clinical assistance, which may occur during a severe pandemic, a reporting system for qualitative indicators could be of great value. There is some evidence that qualitative indicators correlate well with quantitative data [24]. WHO currently encourages reporting of these indicators (along with quantitative epidemiological data if available) through the FluID system [25], which is currently in its piloting phase.
Indirect Indicators of Influenza Circulation
Recently, nontraditional approaches to influenza surveillance have proliferated due to pressure to provide more timely information utilizing electronic information sources. These approaches are usually based on the information-seeking behavior (usually through the Internet) and on healthcare utilization. These methods often utilize syndromic surveillance principles [26]. The rationale for several of the indirect systems is that influenza is the only virus that causes such a rapid, recurrent, and pronounced increase in absenteeism rate and demands for health assistance and information.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


