Species
Chromosomal location
Gene symbol
Size
Other symbols
Human
3p24.2
RARB
423 kb
HAP, RRB2, NR1B2, MCOPS12
Mouse
14 A1-A3; 14 7.08 cM
Rarb
164 kb
Hap, Nr1b2, RARbeta2, A830025K23
Chimpanzee
3
RARB
172 kb
–
Chicken
2
RARB
316 kb
RARBETA
Dog
23
RARB
172 kb
–
In human, RARB consists of 8 exons and 7 introns and spans about 423kb in short arm of chromosome 3. It is located in a chromosomal region which is the host of several genes including CFL1P7, RNA5SP126, TOP2B, MIR4442, CRIP1P2, LOC101927874, RNA5SP125, and EIF3KP2, placed in different directions, consonant oropposite, among which CFL1P7 and RNA5SP126 are located within RARB. Neighboring genes may have important associations to RAR-β functions, but little is known about their function until now, as they are mostly pseudogenes. (See Table12.2 for a short description and function of RARB neighboring genes).
Table 12.2
RARB neighboring genes . (Information is derived from National Center for Biotechnology Information (NCBI), Gene (http://www.ncbi.nlm.nih.gov/gene/5915))
Gene symbol | Gene full name | Direction | Gene type | Function |
|---|---|---|---|---|
CFL1P7 | cofilin 1 (non-muscle) pseudogene 7 | Consonant | pseudo | Unknown |
RNA5SP126 | RNA, 5S ribosomal pseudogene 126 | Opposite | pseudo | Unknown |
TOP2B | topoisomerase (DNA) II beta 180kDa | Opposite | Protein coding | Regulatory roles in transcription and replication; increased proliferation in cancer |
MIR4442 | microRNA 4442 | Opposite | miscRNA | Unknown |
CRIP1P2 | cysteine-rich protein 1 (intestinal) pseudogene 2 | Consonant | pseudo | Unknown |
LOC101927874 | uncharacterized LOC101927874 | Consonant | ncRNA | Unknown |
RNA5SP125 | RNA, 5S ribosomal pseudogene 125 | Consonant | pseudo | Unknown |
EIF3KP2 | eukaryotic translation initiation factor 3, subunit K pseudogene 2 | Consonant | pseudo | Unknown |
Transcription from RARB produces 9 transcripts, 5 protein coding and 4 with no protein product, as a result of alternative splicing (Ensembl:ENSG00000077092), and differential promoter usage (Zelent et al. 1991).
12.3 Protein
RARB produces five protein products, as a result of alternative splicing, with the length of 455 amino acid (aa) being the biggest and 336 aa being the smallest one. Different isoforms differ in their N-terminal regions, which leads the generation of pleiotropic effects of retinoids. RAR-β has two conserved domains, including DNA binding domain of nuclear receptors (NR- DBD)-like superfamily, and ligand binding domain of retinoic acid receptor a member of nuclear receptor family (NR- LBD-RAR). NR-DBD is composed of two C4-type zinc fingers, and interacts with a specific DNA site in the upstream region of target genes and control the rate of transcription. NR- LBD-RAR, on the other hand, provides a binding site for retinoic acid. Upon binding of ligand RAR-β other RARs (α and γ) bind to specific RAR response elements (RAREs), composed of tandem 5′-AGGTCA-3′, in the upstream of retinoid target genes. RAR complex recruit the corepressor proteins AMRT or NCoR when the ligand is absent, and therefor leads toward inactivation of corresponding genes (Marchler-Bauer et al. 2011; Marchler-Bauer et al. 2009; Marchler-Bauer and Bryant 2004). These two domains also exist in several other proteins as examples such as Protein NHR-128 isoform b, estrogen receptor (ER)-β, androgen receptor (AR) (Table 12.3).
Table 12.3
Retinoic acid receptor beta homologous proteins*
Species | Protein symbol | Size | Other names |
|---|---|---|---|
Human | RAR-β | 335 aa | Retinoic acid receptor beta RAR-beta RAR-epsilon HBV-activated protein retinoic acid receptor beta 2 retinoic acid receptor beta 4 retinoic acid receptor beta 5 hepatitis B virus activated protein retinoic acid receptor beta variant 1 retinoic acid receptor beta variant 2 retinoic acid receptor, beta polypeptide nuclear receptor subfamily 1 group B member 2 |
Mouse | Rar-β | 455 aa | Retinoic acid receptor beta RAR-beta nuclear receptor subfamily 1 group B member 2 |
Different isoforms of RAR-β are localized in specific subcellular location, including beta-1 and beta-2 in nucleus, and beta-4 in cytoplasm.
12.4 Gene Regulation
RARB is tightly regulated, indicating its critical roles in vital cellular features. The mouse and human RARB promoters are highly homologous, containing conserved consensus TATA box, retinoic acid responsive element (RARE), TPA-responsive element (TRE), and cAMP-responsive element (CRE). It shows that in addition to the regulation by RARs, RARB can be regulated by several other transcription factors in a well-orchestrated manner (Shen et al. 1991; Dey et al. 1994).
It is shown that RARB is also regulated by microRNAs as well as epigenetic based mechanisms. MIR-16 is located in chromosome 3, and encodes miR-16-2 which binds to three sites in RARB 3’ UTR and inhibits its expression. It is expressed in a variety of cancers, and some normal cells, like different types of breast cancer, pancreatic cancer, prostate cancer, ependynoma primary tumors, renal cortex and medulla, and differentiated embryonic stem cells. miR-16-2 also inhibits nuclear casein kinase and cyclin-dependent kinase substrate 1(NUCKS1) (Hu et al. 2011) (miRTarBase). miR-1 and miR-206 are two other important microRNAs that inhibit RARB expression, and play pivotal roles in myogenesis (Goljanek-Whysall et al. 2012). miR-10a, which is a key mediator of pancreatic cancer metastasis, is shown to be effectively inhibited by RAR (Weiss et al. 2009). Epigenetics based regulation of RARB is evidently among crucial mechanisms that are involved in development and diseases (see below).
12.5 Function and Clinical Implication
RAR-β plays important roles in several developmental, physiologic, and pathogenic mechanisms in human. It takes parts in the development of embryonic digestive tract, myogenesis, eye, hindlimb, and nervous system differentiation, and has crucial negative effects on the regulation of cartilage, cell proliferation, transcription initiated by RNA polymerase II, and more importantly regulate apoptosis (Mark et al. 2009; Mendelsohn et al. 1994; Durston et al. 1989; Niederreither et al. 1999; Maden 2007; Zhu et al. 2009).
RAR-β is a tumor suppressor, and its expression is lost in several malignancies. It induces apoptosis, and is responsible for the chemo-preventive and therapeutic effects of anti-cancer drugs. The importance of RAR-β is notable, as its expression is reduced, or even lost, during different kinds of malignancies like breast cancer, and induction of its expression mediates growth arrest and apoptosis in breast tumor cells. It has been shown that RARs, including RAR-β, also controls cell adhesion by controlling the transcription of involved genes (Al Tanoury et al. 2014). Accordingly RAR-β is identified to actively impedes migration of RA associated breast cancer migration by inhibiting several key migratory proteins like moesin, c-Src, and focal adhesion kinase (FAK) (Inés et al. 2014).
12.5.1 Methylation Status and Diseases
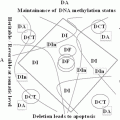
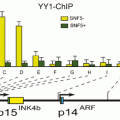
It is demonstrated that RARB5’-region hypermethylation is responsible for suppression of RARB expression in different kinds of cancers (Seewaldt et al. 1995; Liu et al. 1996; Widschwendter et al. 2001; Youssef et al. 2004; Ivanova et al. 2002; Wang et al. 2003; Uray et al. 2009; Twelves et al. 2013; Lotan et al. 1995; Molinari et al. 2013; Cosialls et al. 2012; Chung et al. 2011). It presumably shed light to the important preventive and therapeutic role of retinoids in treatment of cancer. It is investigated that hypermethylated RARB is associated with diagnosis of breast cancer at younger age, with no family history (Pirouzpanah et al. 2010; Mehdipour et al.), high histological grade, high proliferation, increased tumor size, and metastasis (Marzese et al. 2012). In lung cancer, several alterations have been demonstrated in RARB (Gebert et al. 1991). Mutation in RARB is also shown to be associated to microphtalmia (Chitayat et al. 2007; Srour et al. 2013; Chassaing et al. 2013). Recessive and dominant mutations in RARB has shown to cause microphtalmia and diaphragmatic hernia, by loss or gain of function, respectively (Srour et al. 2013). In melanoma, it is shown that RAR-β interacts with p14ARF and plays roles toward the irreversible growth inhibition of malignant cells, therefore treatment of melanoma (Dahl et al. 2013). In Helicobacter Pylori derived gastric cancer, RARB is among genes with reduced expression levels, presumably regulated by epigenetic mechanisms (Cheng et al. 2013). Interestingly among risk factors, benzo(α) pyrene diol epoxide (BPDE) which is a carcinogen present in tobacco and environmental pollution in esophageal cancer, and bile acid which is an oncogene in gastrointestinal cancers, inhibit the expression of RARB (Song et al. 2005; Song and Xu 2001; Li et al. 2002). Methylation pattern of RARB in esophageal squamous cell carcinoma correlates with the development and severity of the disease (Li et al. 2014). Notably, BPDE induces miR-16-2, and as noted above, inhibits RARB (Hu et al. 2011). Unlike, the methylation pattern of RARB is lower in cancer stem cell rich populations of breast tumor (Park et al. 2012), which may play roles in lower proliferation rate of cancer stem cells.
Manipulation of methylation pattern of RARB is getting to be an interesting clinical procedure. Investigating the methylation pattern of RARB has been suggested to be a non-invasive biomarker for the prevention and diagnosis of prostate cancer (Gao et al. 2013). Genetic imbalance in the region containing RARB, on the other hand, has been shown to be a valuable target for personalized medicine (Ribeiro et al. 2014). Interestingly, epigenetic reprogramming seems to have considerable impact on tumor reversion, introducing epigenetic therapy. Axolotl extracts from oocyte reverses the epigenetic silencing of RARB, and therefore tumorigenicity of breast cancer cells, by arresting tumor growth (Allegrucci et al. 2011).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree