Major progress in the understanding of diffuse large B-cell lymphoma (DLBCL) biology has been made in the last decade. Many specific compounds have now entered early phase clinical trials. However, further efforts are needed to find an accurate, fast, reproducible, and affordable technique to translate DLBCL subtype determination by gene expression profiles into clinical application. This article discusses the advantages and drawbacks of the currently available techniques of DLBCL subtype determination as well as important prognostic implications related to the cell of origin. Furthermore, the article provides a schematic description of how molecularly defined DLBCL subtypes could guide tailored therapy.
Key points
- •
Molecular characterization of diffuse large B-cell lymphoma mainly relies on the distinction between germinal center B-cell like and activated B-cell like subtypes.
- •
Each subtype displays specific molecular specificities related to the cell of origin from which they are supposed to derive.
- •
Far from being merely theoretical, the distinction provides important insights into prognostic features, outcome on conventional therapy, and targeted therapy options.
- •
Much work must be performed to implement cell of origin (COO) assessment in routine practice with fast, widely available, and affordable techniques.
Introduction
Diffuse large B-cell lymphoma (DLBCL) accounts for approximately 30% to 40% of all non-Hodgkin lymphoma (NHL) cases. Although the course of the disease is usually aggressive, more than 50% of patients can be cured by currently available immunochemotherapy protocols. The molecular classification of DLBCL segregating germinal center B-cell like (GCB) from activated B-cell like (ABC) subtypes based on gene expression profile analysis remains a cornerstone for the understanding of DLBCL biology. This molecular classification has been proven to be significant for clinical purposes, because it is closely related to patient prognosis. However, major pharmacological progress has only recently rendered specific subtype targeting by new drug compounds possible.
This article focuses on how findings in basic science and translational research have progressively shaped the understanding of DLBCL molecular biology, allowed for better prognostic determination, and uncovered a whole new perspective for treating the disease. It emphasizes the cell of origin (COO) classification of DLBCL, which is supported by many lines of biological evidence and has strong clinical implications. The case of double-hit lymphomas identified by the fluorescence in situ hybridization (FISH) analysis of the rearrangement of MYC and BCL2 or BCL6 oncogenes will be reviewed in another article on this issue.
Introduction
Diffuse large B-cell lymphoma (DLBCL) accounts for approximately 30% to 40% of all non-Hodgkin lymphoma (NHL) cases. Although the course of the disease is usually aggressive, more than 50% of patients can be cured by currently available immunochemotherapy protocols. The molecular classification of DLBCL segregating germinal center B-cell like (GCB) from activated B-cell like (ABC) subtypes based on gene expression profile analysis remains a cornerstone for the understanding of DLBCL biology. This molecular classification has been proven to be significant for clinical purposes, because it is closely related to patient prognosis. However, major pharmacological progress has only recently rendered specific subtype targeting by new drug compounds possible.
This article focuses on how findings in basic science and translational research have progressively shaped the understanding of DLBCL molecular biology, allowed for better prognostic determination, and uncovered a whole new perspective for treating the disease. It emphasizes the cell of origin (COO) classification of DLBCL, which is supported by many lines of biological evidence and has strong clinical implications. The case of double-hit lymphomas identified by the fluorescence in situ hybridization (FISH) analysis of the rearrangement of MYC and BCL2 or BCL6 oncogenes will be reviewed in another article on this issue.
Molecular diffuse large B-cell lymphoma subtypes, prognostic and clinical implications
Why the Need to Classify Diffuse Large B-cell Lymphoma?
The classification of lymphomas is a continuous process that has evolved with the knowledge of the pathophysiology and/or the development of new therapeutics. Since the early recognition of the histologic differences between Hodgkin and non-Hodgkin lymphomas, several classifications have been established. The actual World Health Organization (WHO) 2008 classification recognizes entities based on similar clinical, phenotypical, and/or genetic characteristics. The theoretic framework underlying the WHO classification is the concept of the COO, which proposes to assign every lymphoid cancer to its closest normal counterpart. However, DLBCLs were first considered a highly heterogeneous group of lymphomas that were still too insufficiently characterized to be discriminated.
The ultimate goal of such a classification is to recognize similar lymphomas with the same underlying biological abnormalities so that prognosis and tailored treatment modalities can be chosen. This personalized medicine is often described as the holy grail of modern medicine but is in fact already routinely applied to some lymphoma patients. Hence, therapeutic strategies are already highly differentiated according to the histologic subtype of the aggressive lymphoma (ie, Burkitt lymphoma vs DLBCL). However, many unmet needs remain for a better classification of DLBCL, which represents approximately 40% of adult lymphomas and showing highly heterogeneous clinical outcomes with conventional immunochemotherapy.
Classification of Diffuse Large B-cell Lymphoma Based on Gene Expression Profiles
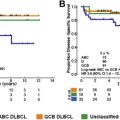
Fifteen years ago, a transcriptomic analysis of fresh frozen samples of DLBCL was reported based on an analysis of approximately 17,000 probes covering genes involved in germinal center biology, lymphoma, cancer biology, and normal lymphocyte biology. The unsupervised analysis of transcriptomic profiling was able to distinguish DLBCL from normal B cells and other lymphoid malignancies. Moreover, this approach clearly distinguished the following 2 classes of DLBCL based on the similarity of their gene expression profiles with normal B cells: The germinal center B-cell like (GCB) and the activated B-cell like (ABC) DLBCL. Importantly, 2 levels of evidence supported the relevance of this distinction. First, the authors noted that specific genetic alterations were associated with each subtype, suggesting distinct oncogenic mechanisms. Second, the GCB/ABC signature was highly correlated with the overall survival (OS) of patients treated with cyclophosphamide, doxorubicin, vincristine, predisolone (CHOP) chemotherapy, and the ABC subtype was associated with the worse outcome.
This pioneering work was further refined 2 years later with a large series of patients with gene expression profiling (GEP) based on 100 genes. This analysis led to the distinction of the following 3 groups: GCB, ABC, and unclassifiable DLBCL. Importantly, this study confirmed the differences in the oncogenetic mechanisms between ABC and GCB DLBCL and the strong prognostic value of this classifier.
Further studies have validated that the worse prognosis associated with the ABC subtype was maintained in the rituximab era, both in frontline therapy and in relapsed DLBCL. Moreover, a predictive value of the COO classification potentially altering therapeutic choice was suggested in relapsed DLBCL patients, because GCB DLBCL was associated with a better survival rate with R-DHAP (containing cisplatin plus cytarabine) than R-ICE (containing carboplatin plus ifosfamide and etoposide). In first-line therapy, young patients with non-GCB DLBCL receiving intensified immunochemotherapy were also reported to experience prolonged survival compared with patients treated with conventional R-CHOP.
Germinal Center B-cell Like and Activated B-cell Like Diffuse Large B-cell Lymphoma: Two Roads to Oncogenesis
Since these 2 early studies, much research has been conducted to analyze the biology of both lymphoma subtypes. Combining whole-exome sequencing and functional unbiased screening with shRNA libraries, it was demonstrated that ABC lymphomas are addicted to the signaling pathways downstream of the B-cell receptor (BCR). This activation of the BCR pathway in ABC DLBCL might be sustained either by mutations in genes encoding this pathway (eg, CD79A , CD79B, or CARD11 ) or BCR activation with self-antigens. Moreover, ABC DLBCLs also frequently harbor an activating mutation of MYD88 , resulting in the activation of the NF-κB pathway and also priming the cells for the detrimental production of interferon β (IFNβ). Interestingly, both pathways have been exploited therapeutically in ABC DLBCL with the use of ibrutinib (which inhibits the Bruton tyrosine kinase [BTK] that belongs to the BCR signaling pathway) and lenalidomide (which favors detrimental IFNβ overproduction in MYD88 -mutated DLBCL by inhibiting interferon responsive factor 4 [IRF4]). In contrast, GCB DLBCLs harbor oncogenetic hits typical of germinal center lymphomas, such as the t(14;18) translocation, the mutations of epigenetic modifiers ( EZH2 , KMT2D ), or mutations in the genes encoding the S1PR2 receptor or its signal transduction protein GNA13 . The main differences in the mutational landscape between the 2 groups are summarized in Fig. 1 .
From Bench to Bedside
The prognostic and predictive potential of DLBCL classification prompted the development of surrogate markers to easily determine the COO. Indeed, GEP using a microarray is not suitable for the clinical routine, because the technique relies on fresh-frozen biopsies, remains expensive when considering numerous samples, and requires time-consuming procedures.
The first attempt to translate the GEP signature of DLBCL for routine purposes was based on immunohistochemistry. One of the most popular algorithms for immuno-histochemistry (IHC)-based determination of COO has been proposed by Hans and colleagues, who described GCB DLBCL as CD10-positive or BCL6-positive and MUM1-negative, while ABC DLBCLs were identified as CD10-negative and MUM1-positive. In an attempt to improve the Hans algorithm, Choi and colleagues (the so-called Choi algorithm), Meyer and colleagues (the Tally algorithm) and Visco and colleagues (the Visco algorithm) proposed different combinations of GC markers (BCL6, CD10, GCET1, and LMO2) and ABC markers (MUM1/IRF4 and FOXP1) to discriminate between DLBCL molecular subtypes. However, there were discrepancies in the clinical outcomes predicted by these different classifiers, and ultimately, none of them was robust enough to classify DLBCL compared with the gold standard GEP classifier. One explanation for these disappointing results was probably the poor reproducibility of the quantitative assessment of the immunostaining. Moreover, these algorithms only recognize 2 categories of lymphoma and fail to identify the unclassifiable cases described in transcriptomic analyses. Nevertheless, this approach is the only one largely available at present, and has been temporarily admitted as an alternative to molecular methods in the recent WHO revision. Although the Hans algorithm is the most popular, this recent revision stipulates that others may also be used.
More recently, RNA-based techniques have been developed that evaluate the level of expression of the most significant genes of the GCB/ABC signature. cDNA-mediated annealing, selection, extension and ligation (DASL, Illumina proprietary technology, San Diego, CA) is a bead-based microarray technology able to quantify mRNA level expression of degraded mRNA from formalin-fixed-paraffin-embedded (FFPE) samples. Although no formal comparison with the GEP gold standard has been performed for ABC, GCB, or unclassified subtype assignments, the technique adequately discriminated patient outcome in a large cohort of 172 patients (of whom 140 were treated with R-CHOP). The technology is currently being evaluated in a randomized clinical trial assessing the addition of bortezomib to an R-CHOP regimen backbone (NCT, ClinicalTrials.gov Identifıer NCT01324596 , REMoDL-B trial).
The NanoString Lymph2Cx 20 genes signature (comprising 15 genes of interest and 5 housekeeping genes) is based on oligonucleotide hybridization of a panel of probes labeled with a fluorescent barcode (digital multiplexed gene expression). For clinical use, this technique can be performed on FFPE samples with a short turnaround time (<48 h). When compared with the gold standard microarray, the Lymph2Cx signature accurately predicted the cell of origin, with less than 1% of cases being misclassified and with a high concordance level between laboratories. Finally, when applied to a large cohort of DLBCL uniformly treated with R-CHOP, the Lymph2Cx signature was able to classify GC versus ABC DLBCL, and the prognosis of ABC DLBCL was poorer as expected. Another study using a 20-gene set that was different from the Lymph2Cx reported the accuracy of the NanoString technology compared with the microarray gold standard. No study has formally compared different gene set NanoString signatures to date. One limitation of these approaches is the need to use the proprietary NanoString platform, which remains expensive for routine use. The feasibility and accuracy of the Lympho2Cx signature in the context of a prospective trial is being assessed (NCT ClinicalTrials.gov Identifıer NCT02285062 , ROBUST trial).
An interesting alternative has recently been developed based on a quantitative RNA analysis called reverse transcription-multiplex ligation-dependent probe amplification (RT-MLPA). Based on the expression level of 14 genes, this technique accurately assigns DLBCL subtype from fresh-frozen or FFPE biopsies, reproducing the prognostic value of the GC/ABC signature. Even if the robustness of this method has been less characterized than the Lymph2Cx signature, its low cost and flexibility support its use as an another approach to assess the COO of DLBCL.
In summary, for 15 years, no reliable technique on FFPE tissue has emerged as a satisfactory surrogate for GEP COO determination. Now that COO determination not only informs patient prognosis but might also drive therapeutic decisions (see the article on targeted therapy elsewhere in this issue), the need for an accurate, broadly accepted, and time- and cost-saving method is more warranted than ever before. Because of its poor reproducibility, there is little chance that immunohistochemistry could become the ultimate surrogate for the GEP gold standard. The NanoString technology seems the most advanced to date in terms of validation against the GEP gold standard and reproducibility between laboratories. However, because of medico-economic considerations, it might be affordable for only a minority of patients. RT-MLPA is an interesting and low-cost technology that has been recognized as an acceptable surrogate in the 2016 WHO classification and warrants further assessment of its interlaboratory reproducibility. Importantly, high-throughput sequencing technologies (next-generation sequencing or NGS) are becoming increasingly available in most care centers and are able to both identify therapeutically actionable target mutations and assess degraded mRNA expression levels from FFPE samples at reasonable costs. One could imagine that a combination of gene expression level determination and targeted sequencing of mutually exclusive mutations (such as GNA13 and MYD88) would become a benchmark in the near future. The pros and cons of the different methods detailed previously are recapitulated in Table 1 .
| Microarray | IHC | Lymp2Cx | DASL | RT-MLPA | |
|---|---|---|---|---|---|
| FFPE compatible | No | Yes | Yes | Yes | Yes |
| Accuracy | Gold standard | Poor | Very high | Undetermined | Very high |
| Reproducibility | High | Low | High | Undetermined | Undetermined |
| Feasibility in daily use | No | Yes | Yes | Yes | Yes |
| Cost | High | Low | High | High | Medium |
Beyond mutation patterns and gene expression profiles, genomic gains and losses encompassing specific oncogenes or tumor suppressors (such as MYC and CDKN1B gains or TP53 and CDKN2A losses) were found to be significantly associated with outcome independently of the ABC and GCB subtypes. As an alternative to genome scale comparative genomic hybridization array (CGHa), multiplex polymerase chain reaction of short fluorescent fragments or quantitative multiplex PCR of short fluorescent fragments (QMPSF) could represent alternatives for routine practice. However, genomic gains and losses remain essentially prognostic and are still lacking any specific therapeutic implications.
Targeted therapy for molecular subtypes of diffuse large B-cell lymphoma
Beyond prognostic issues associated with molecular subtype determination in DLBCL, biological segregation between GCB and ABC lymphomas was found to be of utmost importance for patient-tailored therapy and targeted therapy design. Although promising novel agents in aggressive lymphomas will be detailed elsewhere in this issue, the authors will briefly give an overview of how agents might be particularly attractive for treating specific molecular subtypes of DLBCL (illustrated in Fig. 2 and summarized in Table 2 ).
| Compound | Targets | Rationale | Development Stage | Clinical Trials in DLBCL (Nonexhaustive List) | |
|---|---|---|---|---|---|
| Rationally targeted toward GCB DLBCL | 79–6 | BCL6 |
| Preclinical | None |
| Venetoclax | BCL2 |
| Clinical | NCT01328626 NCT01594229 NCT02265731 NCT02640833 NCT02611323 NCT01969695 NCT02055820 | |
| Obatoclax | BCL2 | Idem | Clinical | NCT00538187 | |
| E7438 | EZH2 |
| Clinical | NCT01897571 | |
| CPI-1205 | EZH2 | Idem | Clinical | NCT02395601 | |
| GSK2816126 | EZH2 | Idem | Clinical | NCT02082977 | |
| Rationally targeted toward ABC DLBCL | Entosplenib | SYK |
| Clinical | NCT01799889 NCT01796470 NCT02457598 NCT02568683 |
| Cerdulatinib | SYK | Idem | Clinical | NCT01994382 | |
| Ibrutinib | BTK |
| Clinical | NCT01569750 NCT01479842 NCT01855750 NCT02142049 NCT02219737 NCT02055924 | |
| ONO-4059 | BTK | Idem | Clinical | NCT01659255 | |
| ACP-196 | BTK | Idem | Clinical | NCT02112526 | |
| Copanlisib | Pan class I PI3K |
| Clinical | NCT01660451 NCT02391116 | |
| Duvelisib | PI3Kγ,δ | Idem | Clinical | NCT01476657 NCT01871675 | |
| ND-2158 or ND-2110 | IRAK4 |
| Preclinical | None | |
| Lenalidomide | Cereblon IRF4 SPIB |
| Clinical | NCT01197560 NCT01122472 NCT02285062 NCT01856192 NCT02128061 | |
| IMO-8400 | TLR7/8/9 |
| Clinical | NCT02252146 | |
| Bortezomib | 26S Proteasome subunit |
| Clinical | NCT02542111 NCT01965977 NCT01324596 NCT01848132 | |
| Ixazomib | 20S Proteasome subunit | Idem | Clinical | NCT02481310 | |
| Carfilzomib | 20S Proteasome subunit | idem | Clinical | NCT01926665 NCT02073097 NCT01959698 | |
| PU-H71 | Hsp90 |
| Clinical | NCT01581541 NCT01393509 |
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree