MicroRNA expression in cancer
Serge Patrick Nana-Sinkam, MD  Mario Acunzo, PhD
Mario Acunzo, PhD  Carlo M. Croce, MD
Carlo M. Croce, MD
Overview
In the last two decades, researchers have identified a novel group of noncoding RNAs (ncRNAs) classified according to their function and size. The best studied of these ncRNAs termed microRNAs (miRNAs) are short noncoding RNAs that are approximately 22 nucleotides in length and play a key role in the regulation of a large number of biological processes and diseases, including cancer. Since the initial description of an association between miRNA and cancer in 2002, miRNAs have emerged as central regulators of processes fundamental to the initiation and progression of cancer. More recently, miRNAs have been detected in bodily fluids including blood, sputum, and urine, thus making them potential noninvasive diagnostic and prognostic biomarkers of disease. The application of miRNAs in human cancer therapy has become a fascinating field of study, representing one of the newest frontiers for cancer treatment. However, our knowledge for these small molecules and their application to human cancers is still growing. In this chapter, we provide an overview of the connection between miRNAs and cancer with a focus on translation to human application.
Background
As investigators increasingly recognize the inherent complexities of cancer, they continue to search for novel molecular pathways in cancer that may be leveraged for the development of novel biomarkers and therapeutics, with the ultimate goal of saving lives. For decades, investigators have held the belief that a large percentage of the human genome was composed of “junk-DNA” or “dark matter” due to its inability to code for proteins. In the last two decades, researchers have now identified functions for regions of the human genome previously considered to be nonfunctional. Some genes located within these regions indeed encode for noncoding RNAs (ncRNAs), with microRNAs (miRNAs) representing the most researched member of this group. Since their initial discovery two decades ago in Caenorhabditis elegans, miRNAs have emerged as key regulators of biological processes fundamental to the initiation and progression of cancers.1, 2 Approximately, 22 nucleotides (nt) in length, miRNAs tend to be highly conserved across species and often demonstrate global deregulation in solid and hematological malignancies.3, 4 By directly binding to either the 3′ or 5′ untranslated region (UTR) of target mRNAs, miRNAs can either degrade target mRNA or inhibit translation. In addition, based on their relatively short size, miRNAs have the capacity for the simultaneous regulation of tens to hundreds of genes, thus interdicting in numerous biological pathways. In fact, the estimate that miRNAs may regulate up to 60% of the human genome is probably wrong and miRNA may regulate over the 90% of the protein-coding genes4, 5). miRNAs are often located in fragile regions of the chromosome and thus susceptible to regulation through chromosomal amplifications, deletions, or rearrangements.6 The mechanisms for the regulation of miRNAs in the setting of cancer are complex and remain only partially understood. However, increasing lines of investigation indicate that miRNA regulation may occur by several mechanisms including alterations in key components of processing, epigenetic silencing, and polymorphisms in either miRNAs or target mRNAs interfering with binding and regulation.7 The mechanisms for miRNA regulation and function are further complicated by their tumor and cell specificity. We are now approaching the identification of nearly 3000 miRNAs. miRNAs may function either as tumor suppressors or oncogenes depending on tumor and cell type and regulate processes fundamental to tumorigenesis (hallmarks of cancer) including differentiation, proliferation, and angiogenesis.8 The mechanisms that regulate miRNAs in cancer, as well as their roles in cancer initiation and progression, are only beginning to be uncovered. While the use of high-throughput profiling strategies for the identification of clinically relevant miRNA-based biomarkers has been useful, this approach to miRNA investigation remains limited by issues of reproducibility and the need for improved algorithms for miRNA-target prediction and validation. Thus, there is still considerable work required to translate miRNAs into markers for clinical decision-making. One must also consider the inherent difficulties in achieving tumor-specific miRNA delivery. As miRNA biology transitions to the clinic, there is encouraging evidence suggesting that human applications for miRNAs, particularly as therapeutics, are in the not-too-distant future. For instance, nanotechnology-based carriers for miRNA delivery represent a new promising tool for the effective shuttling of miRNAs in the human body. Santaris/miRNA Therapeutics Inc. has tested human delivery of an antagomir against miR-122 for the treatment of hepatitis C. More recently, investigators have initiated trials testing human delivery of miR-34 in hepatocellular carcinoma as well as a phase I trial testing miR-16 replacement in recurrent malignant mesothelioma. Such studies represent the first of hopefully a series of future applications for the treatment of human cancer.
Biogenesis and production of microRNAs
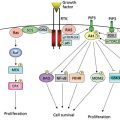
miRNAs are short ncRNAs with a length of approximately 22 nucleotides encoded by evolutionally conserved genes. miRNAs are more frequently located within the introns or exons of protein-coding genes (about the 70%), or in intergenic regions (30%). The expression of intergenic miRNAs is related to their host gene expression, all intragenic miRNAs having independent transcription units.9 miRNAs are processed and generated through a well-orchestrated series of interrelated steps, each of which is currently being investigated (Figure 1). In the first step, a long primary transcript termed the (pri)-miRNA undergoes transcription by RNA polymerase II. The pri-miRNAs are then bound to the double-stranded RNA-binding domain (dsRBD) protein known as DiGeorge syndrome critical region gene 8 (DGCR8) for vertebrates.10 (An RNase III endonuclease termed The Drosha/DGCR8 complex cleaves the pri-miRNA into a smaller stem-loop ∼70-nucleotide (nt) precursor miRNA (pre-miRNA). Pre-miRNAs are then exported from the nucleus to the cytoplasm using the double-stranded RNA-binding protein, Exportin 5 (XPO5).11 Once in the cytoplasm, the pre-miRNA is cleaved into a mature 18–25 nucleotide miRNA by a complex that includes RNAase Dicer, Argonaute 2 (Ago 2), and transactivation-responsive RNA-binding protein (TRBP). The Ago2 protein belongs to the Argonaute family of proteins that bind fragments to guide RNA (including miRNAs). The resultant miRNA sequence, which consists of two strands, is then loaded into the RNA-induced silencing complex (RISC), with the mature strand being maintained while the complementary strand is degraded. The remaining strand possesses complementarity either to the 3′ or 5′ UTR of a target gene leading to degradation through endonuclease activity or to translational inhibition. Recently, investigators have shown that an miRNA can also bind to the coding sequence of a transcript, leading to translational repression.12 The degree of complementarity between the “seed” sequence of the mature miRNA and the target is the primary determinant of the biological effect. It is important to recognize that investigators have now identified additional mechanisms for miRNA biogenesis, suggesting that we are just beginning to understand the complexities of this process.
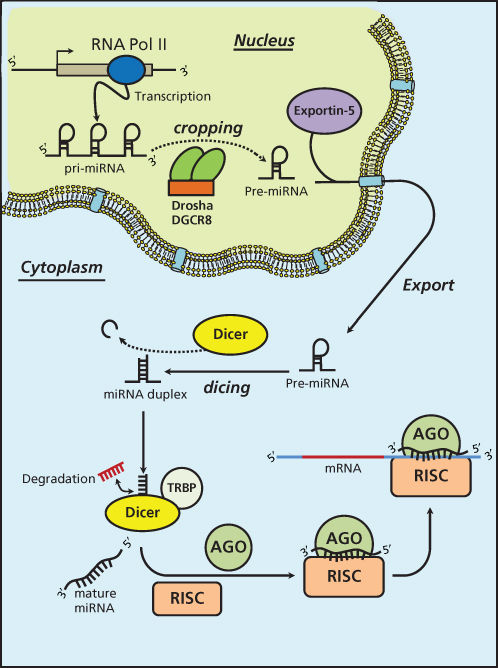
Figure 1 MicroRNA biogenesis.
MicroRNA deregulation in cancer
Calin et al.13 made the first observations linking miRNA deregulation to cancer in a study. While investigating the mechanisms for chronic lymphocytic leukemia (CLL), this team of investigators made the startling observation that a pair of miRNAs, miR-15a/16-1 which, along with the deleted in leukemia (DLEU) gene, are located at the chromosomal region 13q14.3, was either deleted or downregulated in 68% of patients with CLL.13 In addition, both of these miRNAs were highly expressed in normal CD5+ B cells. Both findings suggested that these miRNAs were in fact important to the pathogenesis of the disease. This initial observation was corroborated by independent investigators who identified a functional link between miR-15a/16-1 and the prosurvival molecule Bcl2. They validated in vitro that miR-15a/16-1 targeted Bcl2 to induce apoptosis. Since this initial discovery of miRNAs in CLL, investigators have observed patterns of global dysregulation of miRNAs across both solid and hematological malignancies. In the last few years, researchers have determined that the causes for such dysregulation are multifactorial. For instance, the altered expression and/or function of the proteins involved in the biogenesis of miRNAs, such as Drosha and Dicer, can lead to an aberrant expression of miRNAs and thus to cancer.14 A decreased expression level of Drosha and Dicer has been found in a high percentage of ovarian cancer patients.15 Moreover, epigenetic changes within miRNA promoters such as changes in their methylation can also induce changes in miRNA expression levels.16 Like other deregulated genes that cover an important role as oncogenes or tumor suppressors, the epigenetic deregulated expression of a single miRNA can be the triggering event for carcinogenesis. One such example involves the intensely studied miR-155 whose dysregulation can induce leukemia in miR-155 transgenic mice.17 The tumor-suppressor miR-127 in primary prostate cancer and bladder tumors causes the upregulation of the proto-oncogene BCL6, which is a direct target of miR-127.18 On the other hand, miRNAs can also act as protagonists in the control of the global cellular methylation status by acting on the enzymes responsible for epigenetic control. For example, the miR-29 family is able to modulate methylation levels by affecting the ex-novo expression of DNA methyltransferases DNMT3a and DNMT3B in lung cancer.16 Another important and complex regulative mechanism of miRNAs is related to the transcriptional control of gene expression.19 The activation of the miR-17/92 cluster induced by the MYC oncogene modulates the anti-apoptotic action of E2F1, thus mediating the MYC proliferative effect.20 Recently, the effect of the membrane tyrosine-kinase receptors on miRNA expression has been studied. For instance, the hepatocyte growth factor receptor c-MET is able, through the transcriptional factor AP1, to induce the expression of the onco-miRNA miR-221/222 cluster, suggesting that the important effect of deregulated c-MET in cancer is at least in part linked to a deregulated miRNA expression pathway.21 Finally, considering that the loss of p53 is one of the most represented genetic abnormalities in cancer, the link between the miR-34a family and p53 is another important example of miRNA transcriptional regulation.22 p53 stimulates the transcription of the miR-34 family, inducing apoptosis and senescence. The loss of p53 function induces the downregulation of the miR-34 family in a very high percentage of ovarian cancer patients with a p53 mutation.23 Hence, the primary theme is that patterns of miRNA expression are globally deregulated in cancer, this event being potentially a cause as well as a consequence of cancer itself. The global deregulation of miRNA in cancer has a dramatic effect on downstream targets of several cellular pathways.
Recently, investigators have also determined that miRNA function may be altered through mutations in target gene seed sequences. Such mutations can render an miRNA incapable of regulating a given mRNA and have been identified as biomarkers of clinical outcome. Mutations in the 3′ UTR have been identified in several solid malignancies including ovarian, lung, breast, and colon cancers. Conversely, SNPs in miRNA gene sequences can change miRNA functions. We know that mRNA functional regulation by miRNAs is highly sensitive to base pair mismatches within nucleotides 2–8 of the miRNA, which have been defined as the seed region.24 Therefore a single point mutation on an miRNA gene or a post-transcriptional modification such as RNA editing, can change the function or modify the targetome of an miRNA.25, 26 For example, it was determined that a single nucleotide polymorphism (SNP) in a let-7 (lethal-7) miRNA complementary site in the KRAS 3′ UTR increases nonsmall-cell lung cancer risk.27 A mutation of let-7 binding site in the Kras 3′ UTR has been detected also in juvenile myelomonocytic leukemia (JMML).28
MicroRNA as biomarkers in cancer
Over the last several years, miRNAs have been implicated in virtually every type of cancer. Early studies have focused on applying high-throughput platforms as a means for linking patterns of miRNA deregulation to clinical parameters. One of the first such approaches was made in 2005 by Volinia and colleagues who profiled the miRNA signatures of six human solid tumors, detecting miR-21, miR-17-5p, and miR-191 overexpressed in some tumors.29 Since that initial study, investigators have conducted similar multiple studies with the primary goals of identifying a prognostic miRNA signature. Yanaihara and colleagues conducted high-throughput profiling of cases of stage 1 adenocarcinoma of the lung. They identified over 40 miRNAs that distinguished lung tumors from adjacent uninvolved lung.30 A broader study conducted on 22 different tumor types showed how an miRNA expression profile is able to classify tumors according to tissue of origin with high accuracy.31 Studies have also focused on preselected miRNAs as potential prognostic markers. For example, Nadal et al.32 examined the tumor-suppressive miR-34 as a prognostic biomarker in early-stage adenocarcinoma of the lung. They identified methylation and reduced expression of miR-34b/c in nearly half (46%) of early-stage lung adenocarcinomas and determined that reduced expression and methylation of miR-34b/c correlated with shorter disease-free survival and overall survival. For example, in a separate study, investigators showed that miRNA expression levels may correlate with BCR-ABL kinase activity in chronic myeloid leukemia (CML),33 suggesting, therefore, a potential application in the adjustment of the therapy during treatment to improve the outcome. Another very intriguing application of miRNAs as biomarkers consists of the integration of both protein-coding and noncoding gene expressions in order to develop a prognostic signature in early stages of cancer. Akagi et al.34
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree