At birth, infants become colonized with trillions of commensal microbes that play important roles in health and disease. The majority of these microbes reside in the gastrointestinal tract, reaching densities of 10 12 bacteria per gram of intestinal content in the colon. Complex microbial communities are also found on all human surfaces, including the skin, oropharynx, vagina, and lung. These microbiomes are highly abundant—bacteria represent an astounding 50% to 90% of the cells within or on our bodies and are accompanied by less well-defined but significant numbers of viruses, fungi, and archaea. Overall, the human microbiome contains thousands of species spanning the microbial phylogenetic tree. Because the majority of these microbes are anaerobic and are not routinely recovered using standard microbial culture techniques, the biological impacts of the human microbiome have historically been underappreciated. However, recent technological advances in high-throughput DNA sequencing provided powerful tools to define and study these microbial communities. The microbiome is now understood to be a complex ecosystem in which there is tremendous cross talk between microbial species and between microbes and their host. This communication and interdependence between potential pathogens and the host is likely especially important in severely immunocompromised hosts, such as transplant recipients and oncology patients.
So how does one understand and study the microbiome and its importance to human health? A first step is to catalog the microbes present in a particular anatomic location. Currently, the most widely used culture-independent approach to identify bacterial and fungal microbes is to amplify and sequence the variable regions of the highly conserved 16S ribosomal RNA (rRNA) gene from bacteria or 18S rRNA gene from fungi. These sequences can then be assigned to specific bacteria or fungi and used to determine the abundances of specific microbes within a sample. This approach is inexpensive and high-throughput but has the disadvantage of identifying only bacterial or fungal members of the community. A second approach, referred to as “shotgun” metagenomics, sequences all of the DNA present in a sample. This approach is more expensive and involves substantially more sophisticated analyses to define the members of the microbial community. However, shotgun sequencing provides information on a broader range of microbes, including viruses and archaea, and yields more detailed information on gene content, which can be used to infer the functional capacity of the microbial community.
Taxonomically, microbes are classified by kingdom, phylum, class, order, family, genus, and species. The gut microbiome consists of bacteria primarily from five phyla ( Table 10.1 ). The neonatal gut microbiome has low diversity (relatively few bacterial species) and is typically composed of bacteria from the genera Bifidobacterium and Lactobacillus and facultative anaerobes from the family Enterobacteriaceae. During the first 3 years of life, the gut microbiome is fluid and undergoes substantial shifts in composition with the introduction of solid foods, with strict anaerobes from the orders Clostridiales and Bacteroidales replacing the neonatal microbiome. Alteration of this early life gut microbiome can disrupt immune system development and function. This is most evident in germ-free animals that lack commensal microbes and have myriad health consequences, including abnormal development and function of the immune system and increased susceptibility to infections and autoimmunity. Human epidemiologic studies also reported associations between early life microbiome perturbations (e.g., cesarean delivery, antibiotic exposures) and the later development of asthma, atopy, and autoimmune disorders. Taken together, these studies suggest that microbial exposures are important for educating the developing immune system, with a lack of host-microbe interactions predisposing to immune dysregulation. This concept, often referred to as the “hygiene hypothesis,” was first proposed to explain the geographic distribution of cases of seasonal allergic rhinitis, but has since been extended to other areas of medicine, including hematopoietic stem cell transplantation, oncology, and solid organ transplantation (SOT). ,
| Phyla | Class | Order | Family | Genus |
|---|---|---|---|---|
| Actinobacteria | Actinobacteria | Actinomycetales Bifidobacteriales | Corynebacteriaceae Bifidobacteriaceae | Corynebacterium Bifidobacterium a |
| Bacteroidetes | Bacteroidia | Bacteroidales b | Bacteroidaceae Prevotellaceae Rikenellaceae | Bacteroides Prevotella Alistipes |
| Firmicutes | Clostridia | Clostridiales b | Clostridiaceae Eubacteriaceae Lachnospiraceae Ruminococcaceae | Clostridium Faecalibacterium Eubacterium Blautia Ruminococcus |
| Negativicutes Bacilli | Veillonellales Lactobacillales | Veillonellaceae Enterococcaceae Lactobacillaceae Streptococcaceae | Dialister Enterococcus c Lactobacillus a Streptococcus c | |
| Proteobacteria | Gamma proteobacteria | Enterobacteriales | Enterobacteriaceae a | Enterobacter c Escherichia c Klebsiella c |
| Verrucomicrobia b | Verrucomicrobiae | Verrucomicrobiales | Akkermansiaceae | Akkermansia |
a Bacteria that predominate in infants.
b Bacteria that predominate in children and adults.
c Endogenous bacteria that commonly cause invasive infection.
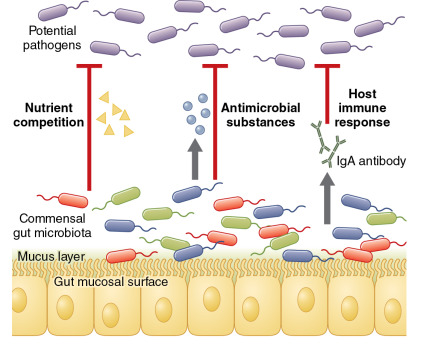
Commensal microbes also prevent infection by providing a barrier to colonization and overgrowth by more virulent bacteria. This concept—referred to as “colonization resistance”—was originally described in 1954 when it was noted that mice treated with the antibiotic streptomycin were susceptible to nontyphoidal Salmonella infection at a dose 10,000-fold lower than the typical minimal infectious dose. The mechanisms that account for colonization resistance are becoming increasingly well understood ( Fig. 10.1 ). It is now recognized that commensal gut bacteria can inhibit pathogen colonization through competition for carbohydrates and other micronutrients, secretion of antimicrobial substances (e.g., peptides, short-chain fatty acids), and through interactions with the host immune system. The classic example of an infection that results from a loss of colonization resistance of the gut microbiome is Clostridium difficile colitis. A loss of gut microbial diversity and anaerobic commensal bacteria, most frequently after the administration of antibiotics, predisposes to C. difficile colonization and infection. Restoration of gut microbial diversity through fecal microbiome transplantation is increasingly being used as a treatment for recurrent C. difficile infection and is widely regarded as the most successful microbiome therapeutic in modern medicine.
Several concepts are important to understanding the impact of the microbiome on human health and disease. First, anatomic location is a critical determinant of the microbiome. For example, the skin microbiome is markedly different from the gut microbiome and, within the gut, the microbiome of the small intestine differs substantially from that of the large intestine. This illustrates the importance of local environmental factors, such as nutrient availability, temperature, and pH, in regulating human microbiome composition. Second, the microbiome performs specific functions, such as digesting dietary fiber, secreting metabolites, and modifying bile acids, and microbiomes with varied composition can provide similar functions. In turn, composition cannot be used to accurately predict microbiome function; microbiomes with similar compositions may perform different functions and have different effects on their host. Third, microbiome alterations observed in the setting of a disease may implicate the microbiome as an important driver of that disease state, or alternatively, may reflect the effect of the disease on the microbiome. Longitudinal studies that include characterization of the microbiome before disease onset are generally needed to establish or refute a causal relationship between the microbiome and a disease state. This is an important distinction to make because it has implications for the potential of microbiome therapeutics to prevent or ameliorate the disease.
In general, microbiomes associated with disease states have lower diversity and high abundances of one or a few potentially pathogenic microbes. This microbiome state is referred to as “dysbiosis” and occurs when the microbiome has a negative impact on the host. A dysbiotic microbiome has disproportionately lost beneficial microbes while microbes that are potentially detrimental to the host have expanded to dominate the microbial community. Such a microbiome may harm the host through loss of microbial metabolic functions that benefit the host. For example, antibiotics that deplete fiber-fermenting anaerobes deprive the host of short-chain fatty acids, which nourish enterocytes and help maintain gut barrier function. Lower abundances of these anaerobes can lead to disruption of the gut mucosal barrier and translocation of intestinal microbes, which can result in bacteremia and sepsis.
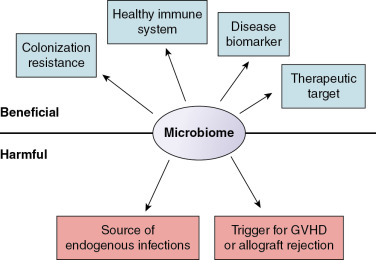
In this chapter, we discuss three emerging themes that relate the microbiome to children undergoing hematopoietic stem cell transplantation (HSCT), treatment for cancer, or SOT ( Fig. 10.2 ). First, the microbiome influences risk for infection. Although many infections in immunocompromised patients originate from endogenous microbes, a healthy microbiome also prevents colonization, overgrowth, and invasion by exogenous pathogens. Second, the microbiome influences immune system function in children at risk for graft-versus-host disease (GVHD) and allograft rejection. Third, the microbiome has the potential to be a powerful tool to predict and prevent infections and other complications in immunocompromised children.
Microbiome and hematopoietic stem cell transplantation
HSCT is associated with substantial alterations of the gut microbiome. Although the gut of healthy individuals typically contains a diverse microbial community composed of approximately 1000 bacterial species, the gut microbiome of patients early after HSCT is frequently far less diverse and is often dominated by a single bacterial species. These shifts in gut microbial diversity and composition occur rapidly—often over the course of days—and are associated with exposure to antibiotics, chemotherapy-induced gut mucosal injury, and dietary changes. Marked increases in the relative abundances of Enterococcus species and Proteobacteria occur frequently after HSCT and are associated with receipt of antibiotics. These shifts are offset by losses of key commensal gut bacteria, including microbes from the genera Faecalibacterium and Ruminococcus . Changes in nutritional intake and, in particular, a lack of enteral intake early after HSCT also contribute to alterations in the gut microbiome. Enteral feeding via a nasogastric tube has been associated with lower GVHD risk and infectious mortality compared with parenteral nutrition in observational studies of allogeneic HSCT recipients. A randomized controlled trial (the NEPHA study) is currently underway to compare these nutritional approaches.
Several studies suggest that the gut microbiome may be a useful biomarker for the prediction of outcomes after allogeneic HSCT. Most of this research has been conducted in adults and has focused on gut microbial diversity. The diversity of the gut microbiome at the time of engraftment is strongly associated with mortality after allogeneic HSCT. In a study of 80 adults, patients with lower diversity of the gut microbiome had markedly worse survival 3 years after allogeneic HSCT (36% vs. 67%). Gut microbial diversity was most strongly associated with mortality from GVHD and infections, suggesting the importance of the gut microbiome to the pathophysiology of these conditions. The gut microbiome also was associated with the risk of relapse among patients undergoing HSCT as treatment for malignancy. In a study of 541 adult patients with hematologic malignancies, the presence of Eubacterium limosum in the gut microbiome predicted a lower risk of relapse at 2 years after allogeneic HSCT.
Graft-versus-host disease
Despite recent advances in histocompatibility matching and donor selection, GVHD remains a leading cause of morbidity and mortality among children after allogeneic HSCT. Our current understanding of the pathogenesis of GVHD suggests that chemotherapy-induced damage to the gut mucosa results in translocation of microorganisms or their products—most notably lipopolysaccharide—triggering an innate immune response that ultimately leads to activation of alloreactive donor T lymphocytes ( Fig. 10.3 ). Research conducted in the 1970s and 1980s established the importance of the gut microbiome to the pathophysiology of GVHD. In these studies, germ-free mice developed GVHD at a far lower rate after allogeneic HSCT than conventionally raised mice. , These findings spurred numerous efforts to prevent GVHD in patients undergoing allogeneic HSCT through suppression or decontamination of the microbiome. A variety of approaches were attempted, including “sterile” diets, laminar airflow isolation, skin cleansing protocols, and gut decontamination through the administration of high-doses of nonabsorbable oral antibiotics. Unfortunately, these strategies were not consistently effective for GVHD prevention in clinical studies and were thus not widely implemented.
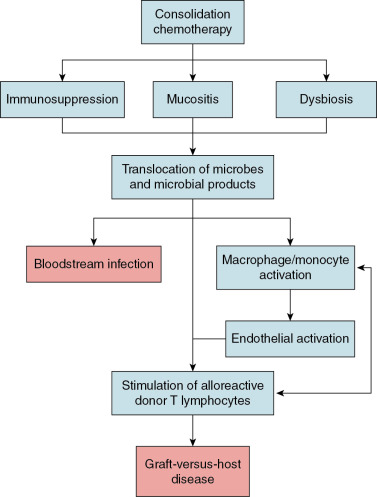
The recent development of high-throughput sequencing technologies led to renewed interest in understanding how the gut microbiome influences the risk of GVHD. Although the diversity of the gut microbiome is a strong predictor of the risk of GVHD, the relative abundances of specific microorganisms also appear to be important. In particular, higher relative abundances of Enterococcus have been observed in patients with GVHD, consistent with studies demonstrating that enterococci can impair gut mucosal integrity and stimulate activation of the innate immune system. Other studies suggest that specific gut anaerobes may be protective for GVHD. In particular, higher relative abundances of certain bacteria from the order Clostridiales (e.g., Blautia ) appear to be associated with a lower risk of GVHD. Limited data from clinical studies also suggest that anaerobic bacteria may be protective from the onset of GVHD. In a single-center retrospective study of adult allogeneic HSCT recipients, the use of antibiotics with an anaerobic spectrum of activity during the peritransplant period was associated with higher GVHD mortality. Although the associations identified in these studies are noteworthy, our understanding of the mechanisms by which the gut microbiome influences GVHD risk remains limited. Further research is needed to better delineate the complex interactions that exist between the gut microbiome and the host immune system to inform strategies to prevent GVHD through manipulation of the gut microbiome.
Infections
There has been much interest in investigating whether the gut microbiome modifies the risk of infections in HSCT recipients. Most studies conducted to date focused on bloodstream infections, particularly those caused by enteric bacteria. Overgrowth of the gut microbiome by Enterococcus or Proteobacteria is associated with a higher risk of bloodstream infection caused by these organisms among adults after allogeneic HSCT. Moreover, antibiotics appear to be an important precipitating factor for overgrowth of the gut microbiome by these bacteria by decreasing colonization resistance. The administration of metronidazole increases the risk of enterococcal domination, consistent with reports that gut anaerobes provide a barrier to colonization and overgrowth by enterococci, whereas fluoroquinolones reduce the incidence of domination by Proteobacteria. Interestingly, and somewhat unexpectedly, gut bacteria also appear to influence the risk of viral infections, including those arising outside the gastrointestinal tract, which could result from the extensive cross talk that occurs between the gut microbiome and the host immune system. Although our understanding of these complex and bidirectional interactions is limited, the gut microbiome appears to play an important role in establishing and maintaining virus-specific memory T-lymphocyte responses. This finding mirrors those of studies of viral infections in germ-free or antibiotic-treated mice in which the presence of commensal microbes alters susceptibility to some viral infections. In clinical studies, the gut microbiome’s influence on viral infections has been most clearly shown for respiratory viruses. Several studies demonstrated associations between antibiotic exposures or the composition of the gut microbiome and the risk and severity of respiratory virus infections among adults after allogeneic HSCT. ,
Microbiome therapeutics
Given the associations between the gut microbiome and the risk of GVHD and infections after HSCT, the potential exists for microbiome therapeutics to improve outcomes of HSCT recipients. These therapies can broadly be divided into four categories: antibiotics, prebiotics, probiotics, and postbiotics. Antibiotics are substances that kill or slow the growth of microorganisms. Prebiotics are nondigestible substances that support the growth of specific microorganisms within the host. Probiotics are supplements or foods that contain live microorganisms. Postbiotics are nonviable microbial products or metabolic by-products that are biologically active within the host.
Antibiotic strategies used in HSCT recipients include antibiotic prophylaxis, de-escalation of antibiotic therapy, and minimizing the duration of antibiotic treatment. Antibiotic prophylaxis is a strategy that is widely used to lower the risk of bacterial infections after HSCT. This practice prevents bloodstream infections and is associated with lower mortality among patients with chemotherapy-induced neutropenia. The most frequent antibiotics used for prophylaxis after HSCT are the fluoroquinolones, most commonly ciprofloxacin. Data from adult HSCT recipients indicate that fluoroquinolone administration prevents overgrowth of Proteobacteria within the gut microbiome, which may provide insight into how these agents reduce bloodstream infection risk. Although antibiotic prophylaxis may be beneficial in HSCT recipients, there is a growing consensus that minimizing disruptions of the gut microbiome after HSCT has the potential to improve patient outcomes. Thus, although patients undergoing HSCT frequently require broad-spectrum antibiotics as empirical therapy for febrile neutropenia, antimicrobial stewardship efforts to limit the duration or spectrum of activity of antibiotics in HSCT recipients should generally be encouraged. As the feasibility of rapidly sampling the microbiome increases in the future, clinicians may be able to serially monitor the gut microbiome to identify patients at high risk of bloodstream infection and to tailor antibiotic therapy to an individual patient’s microbiome.
The administration of probiotics to patients after HSCT is currently an active area of investigation, particularly as a strategy for the prevention of GVHD. Most studies conducted to date have focused on Lactobacillus -based probiotics because of the safety data available for these products in healthy populations. Lactobacillus rhamnosus GG was associated with a lower incidence of GVHD and improved survival when administered to mice after allogeneic HSCT. Moreover, examination of the mesenteric lymph nodes from the mice that received this probiotic demonstrated less translocation of enteric bacteria, suggesting that this treatment improved the integrity of the gut mucosal barrier. Clinical studies of probiotics in HSCT recipients have historically been limited by safety concerns of administering live microorganisms orally in the setting of severe immunosuppression and mucositis. Most notably, there are several well-documented cases of bloodstream infection caused by the microorganisms contained within probiotics among HSCT recipients and other immunocompromised patient populations. However, these complications appear to be rare and there is a growing body of literature to suggest that at least some probiotics may be safe to administer to HSCT recipients. There are currently several clinical trials studying the use of specific probiotics for GVHD prevention in children and adults after allogeneic HSCT. In particular, a double-blind randomized controlled trial of L. plantarum is currently being conducted through the Children’s Oncology Group. This probiotic formulation was previously shown to be safe and well tolerated in a small group of children and adolescents who underwent allogeneic HSCT.
The use of prebiotics or postbiotics is an appealing strategy for manipulating the gut microbiome of HSCT recipients because these products do not include live microorganisms, and thus do not carry a direct infectious risk. There is currently much interest around the use of prebiotic substances to support the growth of Clostridiales and other bacteria that produce short-chain fatty acids. These short-chain fatty acids induce regulatory T lymphocytes that are important for maintaining gut homeostasis and barrier function that may be protective for GVHD. Use of oral β-lactamases is a postbiotic approach that has been proposed to minimize the effect of antibiotic exposures on the gut microbiome. Various compounds are currently in development and several were shown to prevent alterations of the gut microbiome by antibiotics and preserve the colonization resistance provided by commensal gut bacteria in non-HSCT populations.
Fecal microbiome transplantation (FMT) is an alternative to the targeted approaches to modifying the gut microbiome. FMT involves transfer of an entire gut microbial community, rather than one or only a few live bacterial species, and thus has the potential to profoundly alter the recipient’s gut microbiome. Several recent case series reported use of FMT in HSCT recipients. As in other populations, FMT has primarily been used as a treatment for recurrent C. difficile infection in patients after HSCT. In a case series of seven HSCT recipients who underwent healthy donor FMT for this indication, including five patients who were still receiving immunosuppressive therapy, no serious adverse events were reported and only one patient experienced a recurrence of C. difficile . A clinical trial of autologous FMT, in which a fecal sample collected before HSCT is administered back to an individual after HSCT, is currently underway for the prevention of C. difficile infection after allogeneic HSCT. Although the efficacy of this approach remains uncertain, early data from the trial indicate that autologous FMT is effective in restoring microbial diversity to pre-HSCT levels. Finally, based on data implicating the gut microbiome in the pathogenesis of GVHD, healthy donor FMT has occasionally been used with success to treat steroid-refractory or steroid-dependent gut GVHD. However, there is currently little or no experience with the use of FMT during the neutropenic phase that precedes engraftment, and there are concerns that this practice could result in bacterial translocation. Norovirus infection and sepsis have been reported after FMT in other immunocompromised populations, highlighting the need to exercise caution when considering FMT in HSCT recipients.
Microbiome and oncology
We normally co-exist in a healthy equilibrium with our approximately 100 trillion commensal microbes. This balance is maintained by three primary mechanisms: colonization resistance against invasion or overgrowth of pathogens, mucosal barriers, and a robust immune system. These protective mechanisms are frequently lost in children with cancer who are receiving chemotherapy, placing these patients at high risk for serious infections from a broad range of microbes, many of which normally reside at mucosal surfaces. As described earlier, the effect of antibiotics on commensal microbes can lead to a loss of colonization resistance. In addition, there is growing evidence that chemotherapeutic agents may have direct and indirect impacts on the microbiome.
The microbiome drives the development of several types of cancer and also affects clinical responses to cancer treatments. Seminal studies in the 1990s of patients colonized with Helicobacter pylori demonstrated that this bacteria induces chronic gastritis that can lead to gastric cancer, and more recent studies identified associations between the microbiome and colon cancer. Animal and human studies demonstrate that the commensal microbiome influences the efficacy and toxicity of some chemotherapeutic and immunotherapeutic agents, including cyclophosphamide and immune checkpoint inhibitors. The commensal microbiome has also been implicated in the autoimmune complications of immune checkpoint inhibitors; one such case of autoimmune colitis was effectively treated with FMT.
Advances in antimicrobial therapy have improved the outcomes of patients with cancer. In particular, the development of potent antibiotics against Staphylococcus aureus and Pseudomonas aeruginosa in the 1970s (e.g., methicillin and carbenicillin) provided a means to effectively combat active infection by these organisms during periods of neutropenia. More recent data suggest that use of antibacterial prophylaxis can reduce the incidence of infections from chemotherapy-induced febrile neutropenia. For instance, levofloxacin administered during periods of neutropenia prevents bacteremia and decreases the development of febrile neutropenia among pediatric patients receiving chemotherapy for leukemia. However, antibiotics also have a substantial effect on commensal microbes, and a current challenge in the care of pediatric oncology patients is to effectively use antimicrobial agents to prevent and treat infections while minimizing the deleterious effects on the microbiome. A substantial proportion of serious bacterial infections in oncology patients are caused by endogenous microbes from the oropharynx (e.g., Streptococcus species), gastrointestinal tract (e.g., Proteobacteria, enterococci, Candida species), and skin (e.g., Staphylococcus species). High-throughput sequencing technologies have sparked renewed interest in tracking the abundances of these endogenous microbes in oncology patients with the goal of predicting and preventing infections.
Mucosal barrier integrity is maintained by a protective mucus coat, epithelial cell layer, and the mucosal immune system. These components work in concert to prevent translocation of microbes across mucosal surfaces. Chemotherapy-induced mucositis is a major risk factor for bacteremia caused by endogenous microbes such as viridans group streptococci. Although most studies on the pathogenesis of chemotherapy-induced mucositis have focused on the oral mucosa, the same principles are likely to apply to mucositis occurring in other parts of the gastrointestinal tract. In a model promoted by Sonis, chemotherapy initiates free radical generation, inflammation, and epithelial cell apoptosis, leading to disruption of the oral mucosa and translocation of bacteria into the bloodstream. The resulting infections—referred to as mucosal barrier injury bloodstream infections—are especially frequent in children undergoing treatment for acute myelogenous leukemia and solid tumors. Microbes and microbial components translocating across mucosal barriers lead to local and systemic inflammation. This microbial intrusion triggers an inflammatory cascade that changes the local mucosal environment to favor domination by pathogenic bacteria. The importance of mucosal barrier injury in the pathogenesis of bacterial infections in oncology patients has led to the suggestion that “febrile mucositis” might be a more appropriate term to describe oncology patients with febrile neutropenia.
Commensal microbes may also be important for the development of mucositis. In germ-free and selectively colonized mice, the severity of irinotecan-induced mucositis is influenced by the composition of the gut microbiome. Moreover, among pediatric cancer patients, higher baseline oral microbiome diversity was associated with the development of oral mucositis, and children with mucositis had more substantial alterations in oral microbiome composition than children in whom mucositis did not develop. Recent studies suggest that commensal microbes such as Bifidobacterium and Lactobacillus may prevent or ameliorate mucositis by decreasing inflammation and improving epithelial integrity. The microbiome may also indirectly affect infection risk through an influence on hematopoiesis. Germ-free mice display reduced proportions and differentiation potential of specific myeloid precursor cells, and colonization of these mice with a complex microbial community corrects these defects in myelopoiesis. This suggests that the microbiome may facilitate reconstitution of the immune system after chemotherapy in pediatric oncology patients.
Recent studies evaluated the extent to which the fecal and oral microbiomes of pediatric and adult patients are influenced by a leukemia diagnosis. , The largest pediatric study characterized the gut microbiome composition of 199 children receiving treatment for acute lymphoblastic leukemia at St. Jude Children’s Research Hospital. Although the precise impact of cancer treatment on the microbiome requires further study, several themes have emerged. First, patients with cancer undergo a loss of diversity of their oral and gut microbiomes, often associated with domination by microbes, including enterococci, streptococci, and Enterobacteriaceae. Proteobacteria such as Escherichia coli , which typically are of low abundance in the gut microbiome (<0.1%), can expand up to 1000-fold to dominate the gut microbiome. The gains in these potential pathogens are offset by losses of anaerobic bacteria that are important for colonization resistance. These microbiome alterations may result from a variety of factors, including the cancer itself and the associated immune system defects, treatment with antibiotics and chemotherapeutic agents, and the effects of mucositis. Second, alterations in the composition of the microbiome often precede febrile episodes and may be useful in predicting the risk of febrile neutropenia and invasive infections. , For instance, higher abundances of Proteobacteria in the gut microbiome are associated with a higher risk of febrile neutropenia among children with acute lymphoblastic leukemia. Further research is needed to explore the ability of the microbiome to serve as a biomarker for the prediction of infection in pediatric oncology patients.
Treatment of children with cancer with chemotherapeutic and antimicrobial agents leads to alterations of the microbiome, a loss of mucosal barrier integrity, and depletion of the immune system that increase the risk for invasive infection. Antibiotic prophylaxis is an effective strategy for infection prevention in these patients, but this strategy can have detrimental effects on the microbiome that facilitate colonization by exogenous pathogens. Thus the judicious use of antibiotics for the prevention and treatment of infections in this patient population is likely to improve infectious outcomes.
Microbiome and solid organ transplantation
Long-term survival after SOT requires careful manipulation of the immune system to promote tolerance to the allograft while maintaining immunity to protect from infections. This balance is typically achieved through use of immunosuppressive medications. Recently interest has emerged in determining the extent to which commensal microbes affect outcomes after SOT. One hint that commensal microbes may modify the risk of complications after SOT is the higher rates of poor outcomes in patients receiving allografts from sites colonized by commensal microbes (e.g., intestines, lungs) compared with sterile sites (e.g., kidneys, heart), although these mucosal organs are also rich in immune cells and lymphoid tissue. Experimental evidence from germ-free mice demonstrates that commensal microbes indeed influence skin and cardiac allograft rejection. Recent clinical studies indicate that commensal microbial communities change after SOT, with some data suggesting that dysbiosis affects the risk for infection and allograft rejection.
Relatively few pediatric studies have evaluated changes in the microbiome that occur during SOT, but several important concepts are emerging from studies of adults and children. First, similar to allogeneic HSCT, the microbiome is substantially altered by SOT, often characterized by lower diversity and losses of commensal microbes. For example, in a small study of the oral microbiome in pediatric liver transplantation patients, there were alterations in bacterial and fungal microbes in the first few days after liver transplantation. Similarly, cardiac and renal SOT patients also experience microbial dysbiosis, although the relevant anatomic location to study the microbiome in these setting remains unclear. Second, this dysbiosis is influenced by multiple factors, including antibiotic exposures, immunosuppressive medications, anatomic changes (e.g., placement of an ileostomy in intestinal transplantation), and allograft rejection. Third, the influence of immunosuppressive medications on the microbiome varies by agent, dose, and microbiome site. Finally, targeted manipulation of the microbiome is an attractive therapeutic approach to improving outcomes after SOT, and a more complete understanding of the host-microbiome interactions will support microbiome therapeutics that can be used in clinical practice.
Acute rejection of allografts is also associated with microbial dysbiosis. For example, microbial diversity and composition were altered among patients with acute rejection after intestinal transplantation. The gut microbiomes of these patients had lower abundances of Lactobacillales and higher abundances of Proteobacteria. Some studies indicate that reconstitution of the pretransplant lung microbiome among lung transplant recipients protects from bronchiolitis obliterans syndrome, a form of chronic rejection. Shifts in the microbiome also accompany renal transplantation, characterized by losses of diversity and increases in Bacteroidetes and Proteobacteria. , Although it remains challenging to predict how a specific patient’s microbiome will affect immune system responses such as allograft rejection, there are significant potential therapeutic benefits that require further investigation.
Allograft rejection
How might commensal microbes influence the risk of allograft rejection? The intestines and, to a lesser extent, the lungs, harbor large numbers of commensal microbes and receive direct signals from these microbial communities. Although the liver is not thought to be colonized with microbes, it receives portal blood rich in intestinal microbial components. Under certain conditions commensal microbes can translocate from the intestine and be recovered from the liver. As such, intestinal, lung, and liver allografts are anatomically positioned to respond to signals from commensal microbes. In addition, commensal microbes may affect allograft tolerance by regulating activation of the innate and adaptive immune systems. Patients with mutations of toll-like receptor 4 have lower rates of bronchiolitis obliterans syndrome and renal allograft rejection, suggesting that innate immune receptor activation influences allograft rejection. , Finally, in a process referred to as “heterologous immunity,” memory T cells generated from prior infections can cross-react with allograft antigens. Because antigen-specific T cells are also generated against commensal microbes, heterologous immunity may include T cells directed against commensal microbes. Mouse models of allograft rejection demonstrate that the commensal microbiome primes antigen-presenting cells to activate alloreactive T cells, which ultimately leads to allograft rejection. Certain microbes also affect the development of specific subsets of effector T cells, which contribute to alloreactivity. Other microbes, most notably Bacteroides and Clostridium species, promote the development of regulatory T cells that inhibit alloreactivity. Taken together, commensal microbes are critical to the maintenance of balance within the innate and adaptive immune systems, which may in turn influence the risk of allograft rejection.
Microbiome therapeutics
Modulating communities of commensal microbes has the potential to decrease infection and allograft rejection in SOT. Several studies demonstrate that antibiotic-treated or germ-free mice have improved allograft survival. , However, the specific microbes that contribute to allograft rejection have not been identified and antibiotics may have differential effects on the microbiomes of individual patients, which make antibiotics a less appealing approach for manipulating the microbiome of SOT recipients. The administration of probiotics with or without prebiotics in the peritranplant period has also been studied in SOT recipients. A meta-analysis found that probiotics and prebiotics resulted in lower rates of urinary tract and intraabdominal infections in liver transplant recipients but had no effect on allograft rejection or all-cause mortality. Although the results of these early studies are promising, more research is needed to determine the differential benefits that may result from different probiotic strains, dosing, and timing of administration in SOT recipients.
Knowledge limitations
Despite the growing consensus regarding the importance of the gut microbiome to the outcomes of immunocompromised children, there are several limitations to current knowledge on this topic. First, much of the existing clinical data come from single-center studies of adult patients that did not account for potential confounders in microbiome data analyses. This is largely because the statistical approaches for analyzing microbiome data have lagged behind the rapid advances in sequencing technologies that have occurred over the past decade. Microbiome data are highly skewed, sparse, and when collected from the same individual over time, correlated. Currently, there are relatively few statistical methods that can take into account these unique data characteristics, adjust for confounders, and identify the microorganisms that are associated with clinical outcomes. In addition, there are a number of inconsistencies in the findings of studies evaluating the importance of the microbiome in immunocompromised patients. These could be related to differences in practice across centers, further highlighting the need for multi-institutional research, or to variation in the collection, processing, and storage of clinical samples. Finally, the majority of microbiome studies in immunocompromised patients have been conducted in adults. Distinct shifts in the gut microbiome occur during infancy and early childhood, and there are substantial differences in the treatment protocols for HSCT, SOT, and malignancies that are used for children and adults. Pediatric studies are urgently needed to more clearly define how the gut microbiome influences the outcomes of immunocompromised children.
Future directions
Our understanding of the importance of the gut microbiome to immunocompromised children advanced rapidly over the past several decades with the development of high-throughput sequencing technologies. However, the overwhelming majority of these microbiome studies have relied on 16S rRNA gene sequencing, which typically does not have the resolution needed to classify sequencing reads to the species level. Moreover, this approach provides information only on the bacteria that are present in a sample, and specifically does not offer insight into the functional activity of these bacteria or the abundances of fungi, viruses, parasites, or archaea. Increasingly, it is apparent that strain-level differences in genetic content or gene expression can profoundly influence how microorganisms interact with the host. In the future, use of sequencing technologies that can identify microorganisms to the species or strain level and provide information on the functional activity of microorganisms promise to improve our understanding of how the microbiome influences the health of immunocompromised children.
A major goal of microbiome research is to develop therapeutics that can improve patient outcomes. To accomplish this objective, associations observed in clinical datasets must be studied in controlled settings (e.g., in vitro cell cultures, animal models) to confirm the findings and gain insight into potential mechanisms. A powerful approach is to transfer human microbes or microbial communities into germ-free animals to develop gnotobiotic model systems capable of isolating the effect of specific microbes on human diseases. Developing gnotobiotic models of the pediatric microbiome will be a powerful tool to define how microbes interact with the pediatric host. Moreover, although studies of currently available probiotics in immunocompromised children are warranted, the complexity of the gut microbial community and its myriad interactions with the host immune system suggest that a consortium of beneficial microorganisms, selected based on observations in preclinical and clinical studies, may be a promising strategy for modifying the gut microbiome of immunocompromised children.
In summary, commensal microbes have a substantial impact on pediatric HSCT, oncology, and SOT patients. The commensal microbiome modifies the risk of developing certain cancers, the response to and complications from chemotherapy and immunotherapy, and most prominently, the risk of infection and immune dysfunction, including GVHD. A deeper understanding of these dynamic interactions between commensal microbes and the immune system may facilitate the development of microbiome therapeutics that improve outcomes of these vulnerable populations of children.
References
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree