José R. Romero, John F. Modlin
Introduction to the Human Enteroviruses and Parechoviruses
Members of the genera Enterovirus and Parechovirus are Picornaviridae, a large family of morphologically identical, single-stranded, positive-sense RNA viruses that share similar genomic and structural organizations. Originally, the human enteroviruses were divided into five species on the basis of differences in host range and pathogenic potential: polioviruses, group A coxsackieviruses (CV-A), group B coxsackieviruses (CV-B), echoviruses (E), and “newer” enteroviruses (EVs) (Table 172-1).1,2 All enteroviruses discovered since 1970 have been assigned to the enterovirus species.
TABLE 172-1
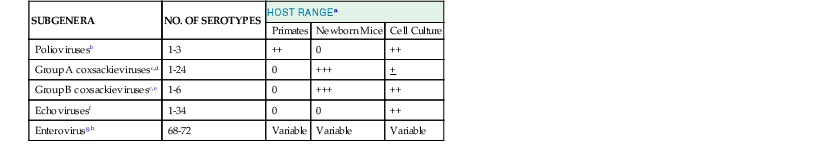
Conventional Classification and Host Range of Human Enteroviruses
| SUBGENERA | NO. OF SEROTYPES | HOST RANGEa | ||
| Primates | Newborn Mice | Cell Culture | ||
| Poliovirusesb | 1-3 | ++ | 0 | ++ |
| Group A coxsackievirusesc,d | 1-24 | 0 | +++ | + |
| Group B coxsackievirusesc,e | 1-6 | 0 | +++ | ++ |
| Echovirusesf | 1-34 | 0 | 0 | ++ |
| Enterovirusg,h | 68-72 | Variable | Variable | Variable |
a Replicative capacity.
b Polioviruses generally replicate only in primates or primate cell cultures, although rare strains such as the type 2 Lansing strain have been adapted to rodents. Although poliovirus multiplies in the alimentary tract of some nonhuman primates, the hallmark of these viruses is the characteristic histopathologic lesions produced by direct inoculation of the central nervous system.
c The coxsackieviruses were first recovered from the feces of children with poliomyelitis in the town of Coxsackie, New York.135 Unlike polioviruses, they produce paralysis and death in experimentally infected suckling mice.
d All group A coxsackieviruses produce generalized myositis of skeletal muscle and flaccid hind limb paralysis in suckling mice,136 and coxsackievirus A7 is pathogenic for the primate central nervous system. However, most group A coxsackieviruses, except serotypes A9 and A16, grow poorly in cell culture. Coxsackievirus A23 has been reclassified as echovirus 9, leaving 23 coxsackievirus serotypes.
e The group B coxsackieviruses are distinguished by their ability to produce focal myositis and generalized infection of the myocardium, brown fat, pancreas, and central nervous system in suckling mice, resulting in spastic paralysis. The group B coxsackieviruses are commonly isolated in cultured primate cells.
f The echoviruses (enteric cytopathic human orphan) viruses were originally discovered in fecal specimens of healthy children.137,138 They cause cytopathic effects in primate cell culture but are generally nonpathogenic for suckling mice (except for echovirus 21) and primates. Echovirus 10 has been reclassified as reovirus 1, and echovirus 28 as rhinovirus 1. Echovirus 34 is a variant of coxsackievirus A24. Echoviruses 22 and 23 have been assigned to the genus Parechovirus as parechovirus serotypes 1 and 2, respectively.82 Therefore, a total of 29 of the original 34 serotypes of echovirus remain.
A classification scheme based on RNA homology within the VP1 capsid protein coding region that replaces the traditional classification divides the genus Enterovirus into four species, designated enterovirus A through D (Table 172-2).2,2a,3 Isolates of the same serotype characteristically diverge in the VP1 region by less than 25% within corresponding nucleotide sequences and by less than 12% within amino-acid sequences.3 Of the original 72 serotypes identified, 64 remain after recognition of redundant serotypes and reclassification of others (see Table 172-1).2 Today, more than 100 unique enterovirus “serotypes” have been identified, each distinguished from one another on the basis of phylogeny of the VP1 coding region.3 Parenthetically, the rhinoviruses, once a genus unto themselves, have been reassigned as species within the genus Enterovirus (see Chapter 177).2
TABLE 172-2
Classification of the Human Enteroviruses by Partial Sequencing of VP1
| SPECIES2a | SEROTYPES |
| Enterovirus A | CV-A2, CV-A3, CV-A4, CV-A5, CV-A6, CV-A7, CV-A8, CV-A10, CV-A12, CV-A14, CV-A16 |
| EV-A71, EV-A76, EV-A89, EV-A90, EV-A91, EV-A92, EV-A114 | |
| Enterovirus B | CV-A9, CV-B1, CV-B2, CV-B3, CV-B4, CV-B5, CV-B6 |
| E-1, E-2, E-3, E-4, E-5, E-6, E-7, E-9, E-11, E-12, E-13, E-14, E-15, E-16, E-17, E-18, E-19, E-20, E-21, E-24, E-25, E-26, E-27, E-29, E-30, E-31, E-32, E-33 | |
| EV-B69, EV-B73, EV-B74, EV-B75, EV-B77, EV-B78, EV-B79, EV-B80, EV-B81, EV-B82, EV-B83, EV-B84, EV-B85, EV-B86, EV-B87, EV-B88, EV-B93, EV-B97, EV-B98, EV-B100, EV-B101, EV-B106, EV-B107, EV-B110 | |
| Enterovirus C | PV-1, PV-2, PV-3 |
| CV-A1, CV-A11, CV-A13, CV-A17, CV-A19, CV-A20, CV-A21, CV-A22, CV-A24 | |
| EV-C95, EV-C96, EV-C99, EV-C102, EV-C104, EV-C105, EV-C109, EV-C113, EV-C116 | |
| Enterovirus D | EV-D68, EV-D70, EV-D94, EV-D111 |
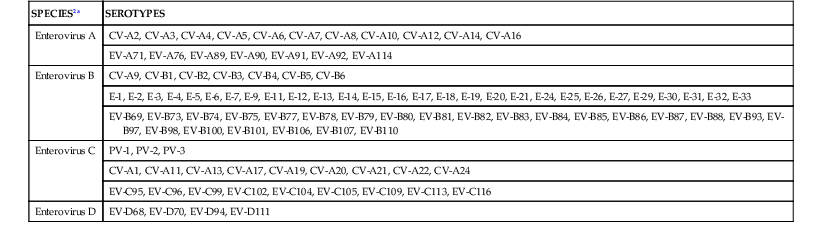
CV-A, group A coxsackievirus; CV-B, group B coxsackievirus; E, echovirus; EV, enterovirus; PV, poliovirus.
Modified from King AMQ, Brown F, Christian P, et al. Picornaviridae. In: King AMQ, Adams MJ, Carstens EB, et al., editors. Ninth Report of the International Committee on Taxonomy of Viruses. New York: Academic Press; 2012:855.
The parechoviruses are members of a more recently established genus that includes two species: Ljungan virus (a virus of rodents) and the human parechoviruses.4 Two serotypes formerly considered to be in the echovirus species, E-22 and E-23, have been reclassified as members of the Parechovirus genus, human parechovirus 1 (HPeV1) and HPeV2,5 on the basis of certain biophysical and biochemical properties that differ from those of enteroviruses (see later discussion). In addition, at least 14 new serotypes of HPeV have been described and are discussed in Chapter 175.
Enteroviruses
Virology
Physical Characteristics
Enterovirus virions are nonenveloped, icosahedral capsids of approximately 30 nm, composed of 60 structural subunits; the subunits are formed from four polypeptides, with an aggregate molecular weight of 80 to 140 kDa, that surround the single-stranded RNA genome.6 Unlike other picornaviruses (e.g., rhinoviruses), the human enteroviruses are stable over a wide range of pH (3 to 10), permitting them to retain infectivity during transit through the gastrointestinal tract. Lacking a lipid envelope, enteroviruses are resistant to ether, chloroform, and alcohol. However, they are readily inactivated by ionizing radiation, formaldehyde, or phenol.6 Molar MgCl2 reduces the thermolability of enteroviruses across a wide range of temperatures; this feature allows live-attenuated oral poliomyelitis (OPV) vaccines to maintain potency when refrigeration is suboptimal or unavailable.
The capsid encloses a linear RNA genome of approximately 7.5 kilobases that is divided into four regions: a 5′ nontranslated region of approximately 740 nucleotides, a continuous-coding region of approximately 6625 nucleotides, a 3′ nontranslated region of approximately 70 nucleotides, and a 3′ poly(A) tail of 70 to 100 nucleotides in length.7,8 The 5′ terminus is covalently linked to a small virus-coded protein (VPg), which is required for the initiation of RNA synthesis. The 5′ nontranslated region contains genomic elements and sequences essential for viral RNA replication, translation and, for some enteroviruses, virulence. The most conserved region of the genome among the human enteroviruses is the 5′ nontranslated region. The regions coding for the structural proteins show the greatest intraserotypic sequence variability, in particular, within the coding regions for epitopes that bind neutralizing antibody. The 3′ nontranslated region plays a role in viral RNA replication. Removal of the poly(A) 3′ terminus renders the RNA noninfectious.
Molecular Biology
The RNA genomes of naturally occurring polioviruses, attenuated polioviruses, and many nonpolio enteroviruses have been fully sequenced, and the replication of polioviruses in primates and transgenic mice has been studied in extensive detail.7,8 Full-length genomic clones of the polioviruses and several enteroviruses have provided the tools for the dissection of many of the molecular events of infection.7–9 The molecular structure and intracellular replicative events appear to be similar for all the enteroviruses.
Host cell susceptibility to enteroviral infection is defined, in large part, by the presence of specific membrane receptor proteins that serve as cellular receptors or co-receptors that bind enteroviruses generally along taxonomic lines (Table 172-3).10,11 The three poliovirus serotypes share a common receptor (PVR or CD155), a member of the immunoglobulin superfamily that is coded on human chromosome 19.11–14 The receptor and co-receptor proteins have been identified for many of the human enteroviruses. Both decay-accelerating factor (DAF, or CD55), a complement regulatory protein, and intercellular adhesion molecule 1 (ICAM-1) play a role in CV-A21 cell entry.15 The CV-Bs also interact with two different cell membrane proteins, the 49-kDa coxsackievirus-adenovirus receptor (CAR) and DAF.16,17 The presence of CAR permits binding and cell entry by all six CV-B serotypes,17 whereas antibodies to DAF block binding and infection by serotypes 1, 3, and 5.16,18,19 DAF also appears to be a major echovirus receptor, binding many E serotypes,20 whereas echovirus serotypes 1 and 8 bind to the α2-subunit of the very late antigen (VLA) integrin molecule.21,22 P-selectin glycoprotein ligand 1 (PSGL-1) and the scavenger receptor class B member 2 (SCARB2) are both used as receptors for EV-A71.23,24
TABLE 172-3
Enterovirus and Parechovirus Cell Membrane Receptors
| RECEPTOR PROTEIN | |
| Enteroviruses (Serotype) | |
| Polioviruses 1-3 | Poliovirus receptor (PVR) |
| Coxsackieviruses A13, A18, A21 | Intercellular adhesion molecule 1 (ICAM-1) |
| Coxsackieviruses B1-B6 | Coxsackie-adenovirus receptor (CAR) |
| Coxsackieviruses B1, B3, B5 | Decay accelerating factor (DAF) |
| Echoviruses 1, 8 | Very late antigen 2 (α2β1) |
| Echoviruses 6, 7, 11, 12, 13, 20, 21, 29, 33 | DAF |
| Enterovirus C70 | DAF |
| Enterovirus A71 | P-selectin glycoprotein ligand 1 (PSGL-1) and scavenger receptor class B member 2 (SCARB2) |
| Parechoviruses | |
| Human parechovirus 1 | αvβ1, αvβ3, integrins |
The processes of penetration, uncoating, and release of the nucleic acid into the cytoplasm occur within minutes at 37° C. Binding of poliovirus with its cellular receptor leads to conformational changes in the viral capsid that allow for extrusion of VP4 and the formation of a channel through which the RNA genome enters the cytoplasm of the cell.8 RNA synthesis begins within 30 minutes, leading to an exponential increase of minus-strand complementary and plus-strand progeny RNA until 2.5 hours after infection, when there is a switch to a linear accumulation of mainly progeny RNA.8 The full-length RNA functions as a monocistronic messenger whose translational product, a polyprotein of molecular weight of 250 kDa, is encoded by a single open reading frame involving about 90% of the entire genome. By convention, the enteroviral polypeptide is organized into three large regions, organized 5′ to 3′ as P1, P2, and P3.25 The nascent polypeptide is cleaved as it emerges from the host ribosome by viral proteins acting initially in cis and later in trans to yield the proteins, and protein intermediates are essential to the viral life cycle.8,25 Cleavage of the P2 and P3 regions results in eight nonstructural proteins and several protein intermediates, whose known functions include polymerase activity, proteolytic cleavage of the translational products, inhibition of host cell protein synthesis, and cellular remodeling.8
Following cleavage of P1 from the nascent polypeptide, it undergoes further cleavages to form the viral capsid, which proceeds by aggregation of five copies each of VP1, VP3, and VP0 (the precursor of VP2 and VP4) into subunits and assembly of 12 of these pentamers into the complete dodecahedral capsid shell. Encapsidation of the viral RNA is associated with a final cleavage of the VP0 protein to VP2 and VP4. The latter is an internal protein closely associated with the RNA. The complete virion contains 60 copies of each of the four structural proteins.8
Host protein and RNA synthesis are severely compromised by 3 hours after infection. After about 6 to 7 hours, virions are visible by electron microscopy within the cytoplasm, and they are subsequently released by lysis of the cell, resulting in a yield of 104 to 105 virions per cell. The number of infectious virions is 10- to 1000-fold lower.8
Pathogenesis and Immunity in Enteroviral Infections
Pathogenesis
The pathogenesis of poliovirus infection has been extensively investigated in primates experimentally infected with neurovirulent strains and in humans infected with vaccine strains,26–29 and it is widely assumed that the early pathophysiologic events of nonpolio enterovirus infections are similar. Studies of coxsackievirus infection in mice have produced much information about the influence of various host and environmental factors on the ability of the virus to replicate in the heart, brain, and other organs and about the mechanism of vertical transmission of enteroviruses from infected pregnant animals to their offspring.
Enteroviruses infect humans via direct or indirect contact with virus shed from the gastrointestinal tract or upper respiratory tract. Whereas ingested virus implants and replicates in the pharynx and the distal small bowel, volunteer studies have shown that attenuated polioviruses replicate most efficiently in the distal small intestine.26 The precise site of viral entry has long been the subject of conjecture. Studies have demonstrated that microfold cells (M cells) expressing the PVR serve to transport polioviruses across the intestinal mucosa.30,31 Enteroviral replication in ileal lymphoid tissue is detectable 1 to 3 days after the ingestion of virus. The quantity of virus recoverable from the tonsils is much less than that in Peyer’s patches, where it may reach 107 to 108 tissue culture median infective doses (TCID50) per gram. In healthy individuals, the duration of viral excretion is typically less than 3 to 4 weeks from the pharynx and 5 to 6 weeks in the feces. Longer periods are reported in persons with B-cell immunodeficiencies and, occasionally, in healthy children.32,33
After multiplication in submucosal lymphatic tissues, enteroviruses pass to regional lymph nodes and give rise to a transient “minor viremia.” This leads to infection and viral replication in reticuloendothelial tissue including liver, spleen, and bone marrow. The most common result is a subclinical infection, in which viral replication is contained by host defense mechanisms. In a minority of infected persons, however, further replication of virus occurs in these reticuloendothelial sites, leading to a sustained “major” viremia that coincides with the onset of the “minor illness” of poliomyelitis and probably of the nonspecific febrile illnesses associated with other enterovirus infections. Prodromal viremia has been demonstrated with wild strains of poliovirus29,34 and E-935 but is uncommon with Sabin OPV strains except for type 2.36
The major viremia results in dissemination to target organs such as the central nervous system, heart, and skin, where tissue necrosis and inflammation occur in proportion to the level of viral replication. Histopathologic lesions are usually not seen in the gastrointestinal tract, even though the small bowel is the site of initial viral replication. The severity of infection in experimental animals can be enhanced by induced exercise, cold exposure, malnutrition, pregnancy, and immune suppression with corticosteroids or radiation.
Viral Mutation during Natural Infection
Enteroviruses undergo a high rate of mutation during replication in the human gastrointestinal tract, and transcription errors occur with a frequency of 1 per 104 bases, approximately one error per genome. As a result, single-site mutations are commonly observed in the 5′ noncoding region of attenuated polioviruses within days after feeding to young infants and such changes are associated with longer excretion and increased neurovirulence.37,38 Serial isolates of the same enterovirus serotype excreted over many years by patients with B-cell immunodeficiency syndromes represent a heterogenetic “swarm” with continuous genetic variation, which can be characterized by oligonucleotide fingerprinting39,40 and RNA sequencing41 and permits estimation of the duration of infection. In addition, RNA sequencing has led to the discovery of virulent circulating vaccine-derived polioviruses (cVDPVs) that have caused outbreaks of paralytic disease among underimmunized populations in a number of countries previously free of poliomyelitis.42–45
Dual infection with different enteroviral strains may produce recombinant progeny virus if the parent strains are in the same class.46 Intertypia can be demonstrated in 1 in 104 to 1 in 105 infectious virions in vitro47 and also in the feces of infants fed trivalent OPV. Most cVDPVs isolated to date are OPV viruses that have recombined with the nonpolio serotypes in the enterovirus C species.43
Immunity and the Immune Response
Immunity to enteroviral infections is serotype specific. Antibody-mediated immune mechanisms operate in the alimentary tract to prevent mucosal infection and in the blood to prevent dissemination to target organs. Neutralizing antibodies target epitopes primarily located on VP1 and less so on VP2 and VP3.48 VP1 possesses the largest number of epitopes as a result of being the dominantly exposed viral capsid protein.8 As early as 1 to 3 days after enteroviral challenge, immunoglobulin M (IgM) humoral antibodies are produced; they predominate in serum during the first month and disappear within 2 to 3 months.28 IgG antibody, which is typically detected by 7 to 10 days after infection, is mostly of the IgG1 and IgG3 subtypes.49 Neutralizing IgG antibodies in serum persist for life after natural infection with enteroviruses.
Small concentrations of humoral type-specific neutralizing antibodies prevented poliovirus viremia and paralysis in experimentally infected primates,50 and passive immunity to paralytic disease in humans can be achieved by the administration of immune serum globulin before exposure to neurovirulent polioviruses.51 However, passively administered immune globulin does not modify the outcome of established central nervous system poliovirus disease52; at this late stage of infection, patients have detectable serum antibody. There is no proven role for immune globulin treatment of other systemic enterovirus infections.
IgA antibody appears in nasal and alimentary secretions 2 to 4 weeks after the administration of live-attenuated OPV and persists for at least 15 years.28 However, mucosal immunity is relative: On reexposure to infectious virus, high titers of secretory IgA antibodies prevent or substantially reduce poliovirus shedding, whereas lower titers are associated with more extensive oropharyngeal replication of virus and longer viral shedding.28 The elaboration of virus-specific IgA antibodies by the small intestine depends on local immunocompetent tissues, as was demonstrated in experiments in infants with double-barrel colostomies who were fed live-attenuated poliovirus through the colostomy and generated secretory IgA antibodies only in the distal loop of the colostomy, not in the pharynx or the proximal loop.53 Antibodies are present in the colostrum and milk of immune women who are nursing and may interfere with the replication of OPV given to breastfed neonates.52 Maternal antibodies passively acquired transplacentally or via milk prevent or modify enteroviral infections of early infancy.54,55
Humoral antibodies have an important role in the recovery from enteroviral infection, as evidenced by the development of persistent infections in persons with significant B-cell immunodeficiency.56 However, there is both clinical and laboratory evidence that humoral antibody alone is not sufficient to limit enteroviral replication in vivo. It has been known for many years that macrophage function plays a critical role in viral clearance, and that macrophage ablation enhances the severity of CV-B infections.57 Recent findings in transgenic mice expressing PVR have shown that the host interferon (INF) response inhibits poliovirus replication in extraneural tissues and limits the spread of poliovirus in the host.58 This activity is likely mediated via interaction with Toll-like receptor (TLR) 3 and the Toll-interleukin-1 receptor domain containing adaptor inducing INF-β (TRIF) pathway.59–62 The TLR3-pathway appears to have a central role in the immune response to human enteroviral infections.
Inhibition of T-lymphocyte function has little effect on virus replication in vivo,63 and persons with abnormal cell-mediated immunity are not predisposed to serious or prolonged enterovirus infections unless they have accompanying B-cell dysfunction.
Even though T lymphocytes do not contribute to the inhibition of enteroviral replication, there is evidence that certain immunopathologic events after enterovirus infection are mediated by T-cell activity. In the murine myocarditis model, expression of proinflammatory cytokines and an acute inflammatory infiltration follow peak viral replication,64–66 and induction of natural killer cell activity and T-lymphocyte immune responses contributes to necrosis of infected cardiac myocytes (see Chapter 86).63,67,68 An inflammatory response may persist long after viral replication has ceased, and ongoing cardiac damage may be mediated by virus-induced antibodies against cardiac antigens69 or by cytotoxic T-lymphocyte–mediated myocyte lysis.65,67,70
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree