Insulin Biosynthesis, Processing, and Chemistry
Christopher J. Rhodes
Steven Shoelson
Philippe A. Halban
Insulin, the major hormonal regulator of glucose metabolism, was first isolated from pancreatic tissue in 1921 by Banting and Best (1). Shortly after the discovery of insulin, the impure extract was used experimentally to treat pancreatectomized dogs (2) and patients with diabetes mellitus (3). As soon as it became clear that the hormone was effective, both the amount of extract isolated and the degree of its purity were increased. Insulin was thus made available to patients with diabetes mellitus. The treatment of such patients underwent a remarkable revolution within 1 to 2 years of the initial discovery of insulin. However, many aspects of insulin biochemistry and chemistry were not understood until years later, and some of these topics remain as areas of intense investigation. This chapter will focus on the biosynthesis of insulin, from insulin gene to secretory granule; the chemistry and structure of insulin; and abnormalities in insulin structure that affect its action or biosynthetic processing.
INSULIN PRODUCTION
In mammals, expression of the insulin gene and insulin biosynthesis are restricted to the β-cells of the endocrine pancreas (4), with the possible exception of the yolk sac and fetal liver (5,6). This exquisite tissue selectivity is conferred upon the insulin gene by a complex interplay between upstream cis -elements (short DNA sequences) and cellular trans -acting factors (proteins) that bind to the DNA and regulate expression (refer to Chapter 2 and Chapter 4). The primary function of the pancreatic β-cell is the production, storage, and regulated secretion of insulin. Under normal circumstances, the β-cell maintains a condition such that there is always a readily available pool of insulin that can be rapidly secreted in response to a stimulus, such as an increase in blood glucose level. Any increase in insulin release is compensated for by a corresponding increase in insulin biosynthesis, so that β-cell insulin
stores are constantly maintained. Thus, the biosynthesis and processing of the insulin molecule along the secretory pathway of the β-cell is a highly regulated and dynamic process.
stores are constantly maintained. Thus, the biosynthesis and processing of the insulin molecule along the secretory pathway of the β-cell is a highly regulated and dynamic process.
Translation of Preproinsulin and Conversion to Proinsulin
Although the two-chain structure of insulin had been known since 1955 (7), it was not until 1967 that Steiner et al. (8,9) first presented evidence for a single-chain biosynthetic precursor, proinsulin. The identification of a hormone precursor was profound for our understanding not only of insulin synthesis but also of protein synthesis in general. It is now evident that there is also a protein precursor for proinsulin, preproinsulin (10,11) (Fig. 5.1), with (for the human protein) the first 24 amino acids forming a signal peptide. The role of signal peptides in the translocation of nascent proteins to the lumen of the rough endoplasmic reticulum (RER) was first elucidated by Gunter Blobel and colleagues (12), and he was awarded the Nobel Prize for this work in 1999. As preproinsulin is synthesized, the signal peptide is bound by the signal-recognition particle (SRP), which in turn homes to the SRP receptor on the membrane of the RER (13). These events occur after the addition of the first 70 to 80 amino acids to the growing preproinsulin chain (14,15). The signal peptide is rich in hydrophobic residues, facilitating the penetration of the RER membrane. Penetration occurs rapidly, and the rest of the nascent preproinsulin molecule then crosses into the lumen of the RER. A signal peptidase within the RER causes cleavage of the signal peptide from preproinsulin to yield intact proinsulin. This proteolytic event arises for the most part cotranslationally, so that preproinsulin is a very minor constituent of the β-cell (16).
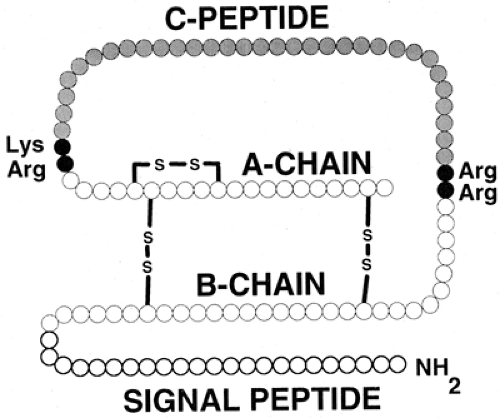 Figure 5.1. Preproinsulin. The initial, high-molecular-weight precursor of insulin consists of four distinct domains. The signal (or pre-) peptide occupies the first (amino-terminal) 24 residues (human preproinsulin) and is cleaved off within the lumen of the rough endoplasmic reticulum to produce proinsulin. Conversion of proinsulin to insulin arises at the pairs of basic residues linking the two insulin (A- and B-) chains to the C- (connecting) peptide, as depicted in Figure 5.3. The two insulin chains are joined together by two disulfide bridges. The molecular mass of insulin is approximately 5,800 daltons. |
Regulation of Insulin Biosynthesis in the β-Cell
The rate of proinsulin biosynthesis is controlled by many factors, including nutrients, neurotransmitters, and hormones (17,18,19,20). The most physiologically relevant of these is glucose (21). The initial studies on the regulation of biosynthesis were limited to measuring the rate of incorporation of labeled amino acid into insulin or its precursors. The earliest of these studies, dating from the 1960s (22,23,24,25,26), showed that glucose specifically stimulated (pro)insulin biosynthesis in vitro above that of general protein synthesis in the β-cell. As for insulin secretion, the metabolism of glucose is necessary to generate intracellular signals to stimulate insulin biosynthesis (21,26,27,28,29). However, while the threshold concentration of glucose required to stimulate insulin secretion is 4 to 6 mM, that required to stimulate insulin biosynthesis is lower—between 2 and 4 mM (28,29,30,31). Maximum proinsulin biosynthesis is reached at a glucose concentration of 10 to 12 mM (21,28,29). As well as glucose (21,30,32,33,34), other sugars (21,23,27,35) and nutrients, including leucine (36), succinate (37), pyruvate (38), inosine, guanosine, adenosine, and ribose (30,39,40), are capable of stimulating insulin biosynthesis but are generally less effective than glucose. After exposure of pancreatic islet β-cells to a glucose stimulus, there is about a 20-minute lag period before there is any significant increase in proinsulin biosynthesis. By 60 minutes, the rate of proinsulin synthesis is stimulated 10- to 20-fold (28,29,41). Upon removal of the stimulus, downregulation of proinsulin biosynthesis is relatively slow, taking more than 1 hour to return to a basal rate (41).
Certain hormones also specifically stimulate insulin biosynthesis, such as growth hormone (42,43), glucagon (44), and glucagon-like peptide-1 (GLP-1) (45). Glucagon and GLP-1 stimulate insulin biosynthesis via a cyclic adenosine monophosphate (cAMP)-dependent pathway; and indeed, cAMP itself can potentiate glucose-induced proinsulin biosynthesis (30,31,32,44,46,47). As for insulin secretion, epinephrine inhibits glucose-induced insulin synthesis (46). Whereas somatostatin is a potent inhibitor of insulin secretion, it has no effect on proinsulin biosynthesis (48,49). Relevant to cytokine-mediated β-cell destruction in type 1 diabetes mellitus, it has been shown that interferons can inhibit insulin biosynthesis, but only at extremely high concentrations (50,51,52). Depending on the concentration used, interleukin-1 can either stimulate or inhibit insulin biosynthesis (52,53,54). The rate of proinsulin biosynthesis is also increased in pregnancy (55,56) and obesity (57,58) as well as during in vivo glucose infusions (59,60,61), but in these examples, it is in association with increased β-cell mass and islet hyperplasia. On the other hand, in other physiologic or pathophysiologic states, such as aging (62), some animal models of type 2 diabetes mellitus (63,64), or during starvation (38,55,65), there is a degree of inhibition of proinsulin synthesis. However, it is not entirely clear whether this is reflective of decreased β-cell mass.
In general, nutrients that are efficiently metabolized in β-cells stimulate insulin secretion and proinsulin biosynthesis at the translational level in parallel (19,20). This provides an ideal situation where insulin released from the β-cell is rapidly replenished at the biosynthetic level by the same stimulus. However, unlike the metabolic stimulus-response coupling for regulation of insulin secretion (refer to Chapter 6 and references 66,67,68,69,70,71), information regarding that for proinsulin biosynthesis is lacking. In all likelihood, beyond a requirement for glucose metabolism, the mechanism responsible for stimulus coupling of proinsulin biosynthesis differs from that for insulin secretion.
There are several lines of evidence to suggest this is so. Extracellular Ca2+ is essential for glucose-induced insulin release, but proinsulin biosynthesis is independent of Ca2+ (66,72). Likewise, inhibitors of Ca2+ calmodulin kinase activity (e.g., trifluoperazine) inhibit insulin release (73,74) but have no effect on proinsulin biosynthesis (75). In contrast, Mg2+ appears to be a requirement of proinsulin biosynthesis, but not necessarily of insulin release (47). Sulfonylureas, which close the β-cell KATP-channel in a key step in signal transduction for glucose-induced insulin release (refer to Chapter 6) (69,71), are very effective at inducing insulin release but appear to have little effect on proinsulin biosynthesis (25,76,77). Likewise, diazoxide that keeps the β-cell KATP-channel open (78) inhibits insulin release (67) but not proinsulin biosynthesis (47). Long-chain fatty acids are marked potentiators of insulin release (79,80,81) but have a tendency to inhibit glucose-induced proinsulin biosynthesis (38,49). Protein kinase C has been involved in stimulus coupling for insulin secretion (82) but does not appear to be related to the specific regulation of proinsulin biosynthesis (38,49,83,84).
There are several lines of evidence to suggest this is so. Extracellular Ca2+ is essential for glucose-induced insulin release, but proinsulin biosynthesis is independent of Ca2+ (66,72). Likewise, inhibitors of Ca2+ calmodulin kinase activity (e.g., trifluoperazine) inhibit insulin release (73,74) but have no effect on proinsulin biosynthesis (75). In contrast, Mg2+ appears to be a requirement of proinsulin biosynthesis, but not necessarily of insulin release (47). Sulfonylureas, which close the β-cell KATP-channel in a key step in signal transduction for glucose-induced insulin release (refer to Chapter 6) (69,71), are very effective at inducing insulin release but appear to have little effect on proinsulin biosynthesis (25,76,77). Likewise, diazoxide that keeps the β-cell KATP-channel open (78) inhibits insulin release (67) but not proinsulin biosynthesis (47). Long-chain fatty acids are marked potentiators of insulin release (79,80,81) but have a tendency to inhibit glucose-induced proinsulin biosynthesis (38,49). Protein kinase C has been involved in stimulus coupling for insulin secretion (82) but does not appear to be related to the specific regulation of proinsulin biosynthesis (38,49,83,84).
In the early 1970s, Permutt and Kipnis convincingly demonstrated that glucose regulation occurred at both the pretranscriptional and posttranscriptional levels (85,86,87) and that translation was affected by modulating initiation and, to a lesser extent, by the rate of elongation (87). Such translational regulation by glucose was confirmed by experiments using cell-free systems (88,89,90). Subsequent studies have found that short-term (less than 4 hours) glucose stimulation of pancreatic islets does not alter the total amount of preproinsulin messenger RNA (mRNA) in a β-cell, even though proinsulin biosynthesis is increased 10- to 20-fold (21,83,91,92,93). Moreover, short-term glucose-induced proinsulin biosynthesis is unaffected by inhibition of insulin gene transcription (28,85). Thus, short-term proinsulin biosynthesis is principally controlled by translational regulation of preexisting preproinsulin mRNA (19,76). Although total preproinsulin mRNA levels in the β-cell are unchanged, upon an increase in glucose level, preproinsulin mRNA is redistributed from a cytosolic storage pool to polysomes associated with the RER, the major site of proinsulin biosynthesis (4,91,93,94). This reaffirmed that glucose primarily influences the initiation phase of preproinsulin mRNA translation (95,96,97,98). The additional glucose-mediated regulation at the elongation phase of translation [below 3 mM glucose; see references (93,99)] and at SRP-SRP-receptor interaction (91,93) is likely adaptive to the primary regulation at the initiation phase of preproinsulin mRNA translation (91,93,100). For general protein synthesis, translational regulation occurs primarily at the initiation phase (98); however, glucose specifically stimulates proinsulin biosynthesis above that of general protein synthesis in the β-cell. Thus, translational control of proinsulin biosynthesis is a relatively exclusive mechanism (21,83,85,100). Although the details of this mechanism have yet to be defined, it is currently thought that there are specific elements within preproinsulin mRNA that control its rate of translation and consequently the biosynthesis of proinsulin (20,100,101). This is analogous to specific translational regulation of ferritin biosynthesis by iron (102).
Under normal physiologic circumstances, where a glucose stimulus lasts no longer than 2 to 3 hours, control of proinsulin biosynthesis is mediated entirely at the translational level (18,28,38,83,93). However, for prolonged exposure to glucose (greater than 4 hours), such as the hyperglycemia in animal models of non-insulin-dependent diabetes mellitus (NIDDM) (103,104), a degree of transcriptional regulation (59,64,105,106) and changes in mRNA stability (107) additionally contribute to the control of proinsulin biosynthesis. During starvation or prolonged hypoglycemia, levels of preproinsulin mRNA are markedly reduced (108,109), in turn reducing proinsulin biosynthesis (55,65). When animals are re-fed after a period of starvation, levels of preproinsulin mRNA rapidly return to normal, allowing proinsulin biosynthesis to recover (108).
Intracellular Trafficking of Proinsulin
From the moment of synthesis, insulin and its precursors, like all secretory proteins, are enveloped in a limiting membrane and as such are protected from the cytosol. The passage from one subcellular compartment to the next occurs by the successive budding and fusion of carrier vesicles, culminating in formation of the mature secretory granule and the ultimate fusion event, exocytosis (Fig. 5.2). These events, initially outlined in pioneering studies by Nobel laureate George Palade (110), provided the point of departure for Orci’s seminal studies on the cell biology of the insulin-producing β-cell (4,111,112). Our understanding of these events, and in particular that of the regulated secretory pathway adopted by insulin and its precursors (113,114,115,116), has progressed, but remains incomplete in its fine molecular detail.
Once in the RER, proinsulin undergoes appropriate folding so that the disulfide linkage between the A- and B-chains of insulin are correctly aligned. Properly folded proinsulin is then delivered to the cis- Golgi apparatus from the RER in transport vesicles in an ATP-dependent process (117,118). The RER to cis- Golgi transport mechanism also requires integrity of the β-cell’s microtubular network (119), Ca2+ (120), guanosine triphosphate (120,121), and certain cytosolic protein factors (120,121,122,123,124,125). Trafficking of proinsulin from the stacks of cis— via the medial— to the trans -Golgi apparatus is mediated by vesicles reversibly encased by members of the COP family of coatomer-proteins (118,126,127,128). Intercisternal transport between Golgi stacks also requires several cytosolic factors and various membrane proteins (118,120,125,126,129,130,131,132,133,134). Once delivered to the trans -Golgi network (TGN), proinsulin is separated from other secretory and integral membrane proteins destined for the constitutive pathway and from enzymes destined for lysosomes (116).
Sorting of Proinsulin to the Regulated Secretory Pathway
Secretion of insulin or other proteins can proceed through either constitutive or regulated pathways (113,135,136). All cells use the constitutive secretory pathway, both for secretion and to dispatch integral membrane proteins to their appropriate destinations. Only a few highly differentiated secretory cell types can store products in secretory granules and release them in response to a secretagogue, however. The β-cell is equipped with both these pathways but under normal circumstances directs proinsulin almost exclusively to the regulated pathway (137). The pattern of release using these two pathways is quite different (113). As its name implies, the constitutive pathway is not subject to control by secretagogues. It arises by the rapid transport of proteins from the trans-Golgi to the plasma membrane in small vesicles (transit time, approximately 10 minutes). For proteins inserted in the vesicle membrane (i.e., cell-surface receptors), fusion of the vesicle with the plasma membrane results in presentation to the extracellular environment, whereas proteins in solution in the transport vesicles are secreted.
The regulated pathway involves the selective packaging of secretory proteins (for the most part in the form of proproteins, and notably prohormones) into secretory granules, followed by
secretion (exocytosis) in response to a stimulus. Active selection of proinsulin destined for granules is known to take place in the trans -most cisternae of the Golgi complex (138). The constitutive pathway, by contrast, is considered to be a default route, accepting any material in the trans -Golgi not previously singled out for shipment to a specific destination. In the rat, the sorting process has been shown to be remarkably effective, with more than 99% of newly synthesized proinsulin entering the regulated secretory pathway (137). If this is the case for primary β-cells, sorting to granules appears to be less efficient in transformed cells (139).
secretion (exocytosis) in response to a stimulus. Active selection of proinsulin destined for granules is known to take place in the trans -most cisternae of the Golgi complex (138). The constitutive pathway, by contrast, is considered to be a default route, accepting any material in the trans -Golgi not previously singled out for shipment to a specific destination. In the rat, the sorting process has been shown to be remarkably effective, with more than 99% of newly synthesized proinsulin entering the regulated secretory pathway (137). If this is the case for primary β-cells, sorting to granules appears to be less efficient in transformed cells (139).
The selection process for directing prohormones to granules is not well understood. There is morphologic evidence for an intimate association of proinsulin with the cisternal face of Golgi membranes, suggestive of the presence of receptors (140). If such receptors exist, their unequivocal identification remains elusive. Thus, the postulated role of carboxypeptidase E (also known as carboxypeptidase H), an enzyme involved in the trimming of prohormones following endoproteolytic cleavage (see below), as such a receptor (141,142,143) has been challenged both in general terms (144) and more specifically in the context of proinsulin sorting (145,146,147). An alternative sorting mechanism implicates selective protein aggregation in the TGN, leading to concentration/condensation in intimate association with the TGN membrane (116,148,149,150). It is proposed in this model mechanism that common physicochemical properties of secretory-granule proteins allow them to aggregate (or coaggregate) in the presence of an elevated Ca2+ concentration and a mildly acidic pH as encountered in the internal environment of the TGN (116,149,150,151,152,153) and that this event in itself promotes budding of a nascent granule. However, sorting by aggregation is likely to be a complex mechanism, considering that 50 to 100 β-granule proteins (154,155) must be coordinately delivered to the site of secretory granule biogenesis (59,154,156).
Regardless of the true TGN sorting mechanism, it is commonly supposed to be similar in all cells equipped with the regulated pathway. This conclusion is based in part upon the results of experiments in which cells are transfected with genes for foreign proteins. For example, AtT20 (pituitary corticotroph) cells can target foreign secretory peptides such as proinsulin (157) or growth hormone (158) as well as exocrine enzymes such as trypsinogen (159) to secretory granules. Furthermore, a protein normally destined for the constitutive pathway can be rerouted to secretory granules and regulated release if fused to the appropriate sequences of the growth-hormone molecule (160). The last experiment indicates that the targeting machinery
must recognize functional domains on the secretory protein. By analogy, there must be functional domains on the proinsulin molecule allowing for its recognition and targeting to the regulated pathway (161). If so, they remain to be identified and characterized experimentally. Although a mutant proinsulin (with aspartate instead of histidine in position 10 of the B-chain) has been shown to be diverted from the regulated to the constitutive pathway (162,163), the reason is not clear. It could be due to perturbation to a “sorting domain,” lack of coordination of Zn2+ needed for hexamerization [unlikely because another mutant proinsulin unable to form hexamers appears well sorted (164)], or to more esoteric reasons related to the enhanced binding of this particular mutant proinsulin to the insulin receptor (163,165).
must recognize functional domains on the secretory protein. By analogy, there must be functional domains on the proinsulin molecule allowing for its recognition and targeting to the regulated pathway (161). If so, they remain to be identified and characterized experimentally. Although a mutant proinsulin (with aspartate instead of histidine in position 10 of the B-chain) has been shown to be diverted from the regulated to the constitutive pathway (162,163), the reason is not clear. It could be due to perturbation to a “sorting domain,” lack of coordination of Zn2+ needed for hexamerization [unlikely because another mutant proinsulin unable to form hexamers appears well sorted (164)], or to more esoteric reasons related to the enhanced binding of this particular mutant proinsulin to the insulin receptor (163,165).
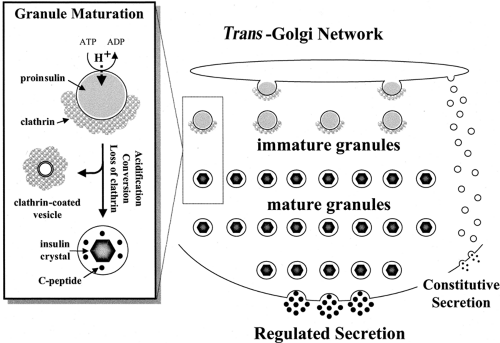
The Clathrin-Coated, Immature Secretory Granule
The earliest detectable form of the secretory granule in β-cells is formed from regions of the trans -Golgi characterized by the presence of clathrin on their cytosolic face (166), and the immature granule similarly carries a partial coating of clathrin (111). Clathrin consists of two chains, which can associate to form a network of triskelions and cover membranes with a basket-like coat (167,168,169). The assembly of a clathrin coat depends on associated proteins, and coating of secretory granules involves the adaptor protein AP-1 (149,170) as well as ADP-ribosylation factor-1 (ARF-1) (170,171). It appears increasingly likely that clathrin is involved in the purging of unwanted proteins from granules during their maturation (see next section) rather than in the formation of the immature granule from the TGN (150).
Granule Maturation and Refinement of Granule Constituents
Maturation of the immature granule involves three parallel events: loss of clathrin, progressive acidification, and proinsulin conversion. The loss of clathrin is attributed to the budding of clathrin-coated vesicles from maturing granules (150), as first proposed by Orci (111). They are understood to take with them some soluble granule proteins, with insulin being retained (“sorting by retention”) in the granule due to its crystalline state (see below) (172,173). This results in the purging of unwanted proteins that may have been erroneously introduced into the immature granule. A second mechanism, “sorting for exit,” operates in parallel to this same end. This involves the selective binding of proteins to the intragranular aspect of mannose-6-phosphate (and possibly other) receptors that participate in coordinating clathrin adaptors via their cytosolic domain. Lysosomal proteases are known to be bound in such a fashion, leading to their sequestration in clathrin-coated vesicles and consequent clearance from maturing granules (150,174,175).
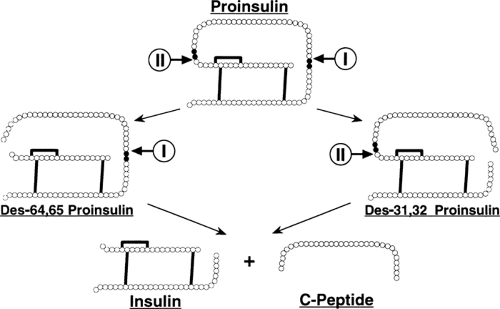
Conversion of Proinsulin to Insulin
The conversion of proinsulin to insulin and C-peptide occurs in the immature, clathrin-coated secretory granule (4,176,177) and is outlined in Figure 5.3. There are two pairs of basic
residues at which the endoproteolytic conversion event occurs: Arg31-Arg32, linking the insulin B-chain to the C-peptide; and Lys64-Arg65, linking the C-peptide to the insulin A-chain.
residues at which the endoproteolytic conversion event occurs: Arg31-Arg32, linking the insulin B-chain to the C-peptide; and Lys64-Arg65, linking the C-peptide to the insulin A-chain.
The Proinsulin-Processing Enzymes
Endoproteolytic peptide-bond cleavage of proinsulin occurs on the carboxyl side of Arg31, Arg32 and Lys64, Arg65, followed by exopeptidic removal of the newly exposed basic amino acids (180,181) (Fig. 5.3). The proinsulin-processing endopeptidase activity proved elusive and was not discovered until 20 years after that of proinsulin (182). Soon after, it was found that this activity actually consisted of two individual endopeptidases, then named type I and type II (183). Type I cleaved specifically at the Arg31, Arg32 site of proinsulin, and type II preferentially cleaved the Lys64, Arg65 site (Fig. 5.3) (183,184,185,186,187). Soon after, these endopeptidase activities were cloned and found to be members of a family of prohormone processing enzymes (188,189,190,191). The type I endopeptidase activity was equivalent to PC3 (also known as PC1) (192,193,194,195), and the type II endopeptidase activity was equivalent to PC2 (152,196,197,198). Cotransfection studies of proinsulin with PC3 and PC2 have confirmed these enzymes as the bona fide proinsulin-processing endopeptidase activities (199).
The third proteolytic enzyme involved in proinsulin processing is an exopeptidase, carboxypeptidase H (CPH) (180,181). CPH [also named CPE (200)] is a Ni2+-requiring exopeptidase with an acidic pH optimum (181). Once proinsulin has been cleaved by PC2 or PC3, CPH specifically and rapidly removes the exposed (C-terminal) basic amino acids (181,183,186).
Regulation of Proinsulin Conversion
PC2 and PC3 are Ca2+-dependent enzyme activities that display an acidic pH optimum of around 5 to 5.5 (183). Maximal activity of these enzymes is appropriately assured by the Ca2+-rich [1–10 mM free Ca2+; see reference (201)] and mildly acidic [pH 5.5; see reference (202)] internal milieu of an immature insulin secretory granule (183,185,194,195), which is also optimal for CPH activity (181). Because these [Ca2+] and acidic pH conditions are not encountered in the secretory pathway of the β-cell until the immature insulin secretory granule compartment is reached, it ensures that insulin is produced only in the intracellular β-granule compartment in which it is stored (183,203). As such, for full PC2, PC3, and CPH activity, activation of the secretory granule proton-pumping ATPase to generate an acidic pH (176,179,202) and Ca2+-translocation proteins to increase [Ca2+] (204) are key regulatory events for controlling proinsulin conversion.
Most members of the proprotein processing endopeptidase family undergo a zymogen-like autocatalytic cleavage activation (205,206,207). In pancreatic islets, PC2 and PC3 are synthesized initially as inactive proproteins that are subsequently cleaved by limited proteolysis (29,197,208,209,210). This processing of PC2 and PC3 precursors is thought to be initiated in the RER (152,211) and then completed in the TGN/immature granule compartment (152,211,212). This also ensures that the vast majority of proinsulin is converted to insulin in the secretory-granule compartment (4,203). While it is likely that proteolysis of proPC2 and proPC3 is an autocatalytic process, processing by other endoproteolytic activities has not been ruled out (152,207,211,212). Recently, it has been found that the proteolytic processing of proPC2 is more complex than originally thought because of the transient interaction between proPC2 and its chaperone, 7B2, that inhibits PC2 activity (213,214). Proteolytic cleavage of 7B2 results in subsequent PC2 activation in the trans -Golgi region of the secretory pathway (215,216). In part, this may account for the observation that PC2 traverses the secretory pathway more slowly than PC3 and proinsulin (152,208).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree