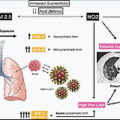
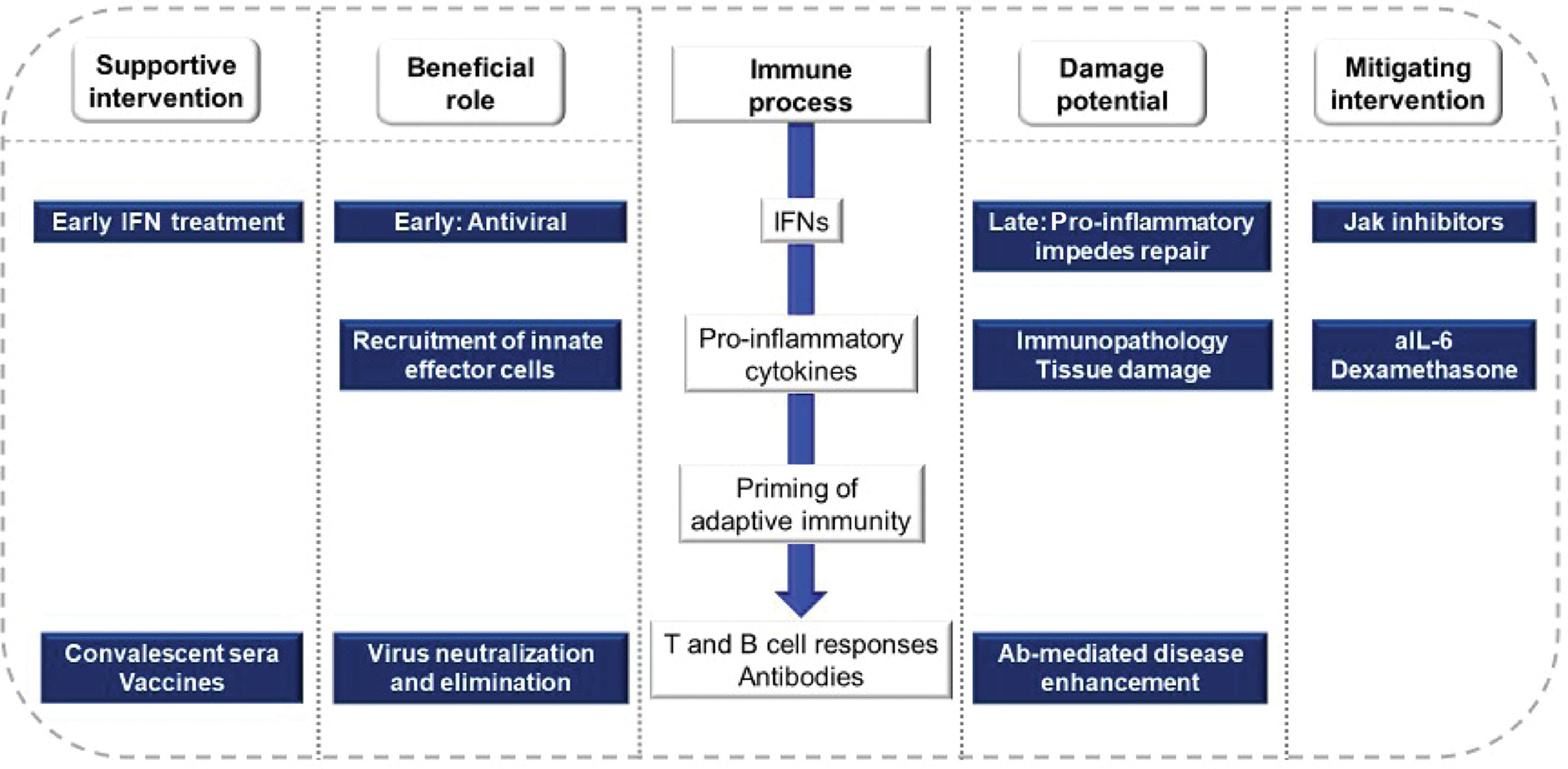
The extraordinary global spread of SARS-CoV-2 through naïve populations provides compelling evidence of both the strengths and limitations of the human immune response: on the one hand, recognizing and eliminating a novel viral pathogen, while on the other, causing potentially lethal immunopathological conditions ( Fig. 3.1 ). The pandemic also provided a striking opportunity to understand SARS-CoV-2 pathogenesis through a lens offered by multiple clinical trials. COVID-19 disease seems best considered in two phases: a brief early phase with high-level viral replication and limited immune responses, and a later phase in which these are reversed. A minority of infected persons progress to life-threatening illness in the later phase of infection, with striking consistency across diverse populations. Except for age, the risk for progression in most individuals cannot yet be attributed to a single critical social, economic, environmental, or genetic factor. For immunologists, the key emerging research questions regard how early responses to SARS-CoV-2 affect subsequent risks for severe disease and the underlying protective and pathogenic mechanisms. To this end, this chapter will discuss innate and adaptive immune responses to SARS-CoV-2 infection, the role of immune dysregulation and its contribution to severe pathological processes, and the potential role of endotypes to guide future studies of immunotherapy.
Innate Immune Responses
Early responses to many infections are driven by activation of cellular pattern recognition receptors (PRRs). Genetic sequences for these receptors are fixed: unlike immunoglobulin or T-cell receptor genes, they do not undergo rearrangement in somatic cells. PRRs located on cell membranes, such as Toll-like receptors (TLRs), are generally triggered by microbial ligands, whereas those in the cytoplasm may be triggered by indicators of molecular damage (damage-associated molecular patterns [DAMPs]) or by RNA or DNA with specific characteristics. As a single-stranded RNA virus, SARS-CoV-2 may be recognized by TLRs 7 or 8 (recognizing single-stranded RNA), retinoic acid–inducible gene I (RIG-Ia) (recognizing uncapped viral RNA), or melanoma differentiation–associated protein 5 (MDA5), an RIG-I–like PRR recognizing long double-stranded cytosolic RNA. PRR triggering results in a signaling cascade that includes production of interferons (IFNs), so named in the 1950s for their ability to interfere with virus propagation ( Fig. 3.2 ). Many IFN-stimulated genes (ISGs) encode proteins with direct antiviral effects ( Fig. 3.3 ). The breadth of ISG responses hampers the emergence of viral avoidance mechanisms, although viruses nonetheless devote substantial genomic space in an attempt to do so. This competition between viruses and host cells is one of the strongest drivers of protein adaptation in mammalian evolution. Emerging evidence indicates that the enhanced transmissibility of new SARS-CoV-2 variants is due in part to enhanced evasion of innate responses.

The IFN-driven innate response to viral infections has three important functions: creating an intracellular milieu that limits viral replication, recruiting immune cells to the site of infection, and priming an adaptive immune response through the expression of costimulatory molecules. However, the very breadth of the innate response can also pose significant costs to the host. This is best illustrated by clinical experience with various forms of recombinant IFN-α, which, for more than a decade, was combined with ribavirin for treatment of chronic hepatitis C virus infection. Together, these drugs could cure about half of patients; factors increasing the likelihood of cure included low pretreatment levels of IFN-induced protein 10 (IP-10). , However, IFN treatments were poorly tolerated, with black box warnings of fatal or life-threatening neuropsychiatric, hematological, autoimmune, ischemic, and infectious disorders. In routine clinical use, less than 25% of patients were able to complete a full course of treatment. The IFN regimens have largely fallen out of use now that alternatives (direct-acting antiviral drugs) are available.
IFN appears essential for protection against SARS-CoV-2, based on reports of life-threatening disease in persons with inborn errors or autoantibodies affecting pathways for type I IFNs (α and β). , Early IFN responses appear insufficient for optimal control of viral replication even in the absence of recognized immune defects. The clearest evidence for this comes from a small trial in which 60 newly diagnosed COVID-19 outpatients were administered a single dose of pegylated IFN-λ (PEG–IFN-λ) or placebo. IFN recipients were more likely to have undetectable virus by day 7 (odds ratio [OR], 4.12; P = .029). The benefit was greatest in those with high viral loads at baseline. Receptors for IFN-λ (a type III IFN) are expressed on epithelial cells but not on lymphocytes, potentially preserving efficacy while increasing safety and tolerability. Further trials comparing subtypes of the IFN-α, β, or λ families are underway to test this hypothesis.
A trial conducted in Hong Kong examined the addition of IFN-β plus ribavirin to lopinavir/ritonavir in 127 patients. Although subjects in this trial were inpatients, its patient population was similar to that of the IFN-λ trial, in that only 13% required supplemental oxygen at baseline. IFN treatment yielded a shorter median time to negative polymerase chain reaction (PCR) (7 vs. 12 days; P = .0010), and a shorter time to resolution of symptoms (4 vs. 8 days; P < .0001). In contrast, the World Health Organization Solidarity Trial examined IFN-β treatment in 243 hospitalized patients, most of whom already required oxygen therapy. In this population with more advanced disease, IFN treatment tended to increase mortality risk compared with controls (relative risk [RR], 1.16; P = .11). Similarly, a retrospective study published early in the pandemic suggested that the likelihood of survival was increased by early IFN-α treatment (within 5 days of hospitalization), whereas survival was reduced if IFN was started later. Several studies have confirmed that IFN levels correlate generally with COVID-19 disease severity; that is, IFN levels are highest in persons with severe disease. One study suggests that mutations outside of the spike coding region contributed to the evolutionary success of SARS-CoV-2 variants (most notably alpha) through the enhanced suppression of innate responses. However, it seems that for later variants, evasion of antibody responses (induced by vaccination or prior infection with early lineage strains) may better account for evolutionary success (discussed below).
Together, these studies support a hypothesis that innate responses during early SARS-CoV-2 infection are insufficient for viral control, and that the subsequent amplification of these responses contributes to immunopathological processes.
Adaptive Immune Responses
The humoral and cellular arms of the adaptive immune system are integrally linked with innate immune mechanisms in the overall response to viral infection. Distinct mechanisms of each system are engaged differently depending on virus type, the timing of a response, and the cell types involved ( Table 3.1 ). Unlike innate responses, which recognize patterns shared by many pathogens, adaptive immune responses recognize specific antigenic sequences of specific pathogens. The required receptor diversity is accomplished by gene rearrangements for antigen receptors or immunoglobulin regions early in the development of T and B cells, respectively. Antigen recognition triggers both cellular activation and clonal expansion. Depending on the frequency of precursor cells and the nature of the stimulus, a period of days to weeks must elapse for the evolution of a primary adaptive immune response.
| Dendritic Cells | Macrophage | B Cell | CD4+ T cell | CD8+ T cell | |
|---|---|---|---|---|---|
| Location | Skin, mucosal epithelium (Langerhans cells), bone marrow, blood, spleen, thymus, tonsil, liver, lung, intestine, lymph nodes | Throughout the body | Blood, peripheral lymphoid organs | Throughout body | Throughout body |
| Function | Antigen-presenting cell of innate immune system Both extracellular and intracellular antigens | Antigen-presenting cell of innate immune system Extracellular antigens , | Adaptive immunity: Antibody production Antigen-presenting cell Effector cell killing of infected cells , | Effector cell terminating infection B-cell help CD8+ cell help Cellular memory Type 1 intracellular and type 2 extracellular (e.g., helminths) response to infections , | Effector cytotoxic killing of infected cells Type 1 response to intracellular infections , |
| Specific antigen receptors | No | No | Surface immunoglobulins | TCR | TCR |
| Differentiation | cDC1 and cDC2 | M1 and M2 | Memory B cells Plasma cells | Tfh-promoting B cell response in lymphoid tissue Treg: Maintenance of immunological tolerance to self and foreign antigen Th1: Eliminates intracellular pathogens and associated with organ-specific autoimmunity Th2: Produces an immune response against extracellular parasites, and play major role in induction and persistence of asthma and other allergic diseases Thl7: Produces immune response against extracellular bacteria and fungi. Also involved in the generation of autoimmune diseases | Tc1: Immune response against intracellular pathogens and tumors Tc2: Th2-mediated allergic reaction, contributes to arthritis Tc9: Inhibits CD4+ T-cell mediated colitis, propagates Th2-mediated allergic reaction, antitumor response Tc17: Propagates autoimmunity, confers immunity to viral infections, antitumor response CD8+ Treg: Regulates T-cell–mediated responses |
| Costimulation/cytokines | ICOS, CD28 | M1 is induced as a response to infectious stimuli (e.g., LPS and/or IFN-γ) and M2 is induced by resolving stimuli (e.g., IL-4) | IL-21, IL-4, CD40L | CD28 | CD28 |
| Interferon production | Type I and type III interferons | Type I interferons | IFN-γ | IFN-γ | Type I interferon protects antiviral CD8+ T cells from NK cell cytotoxicity |
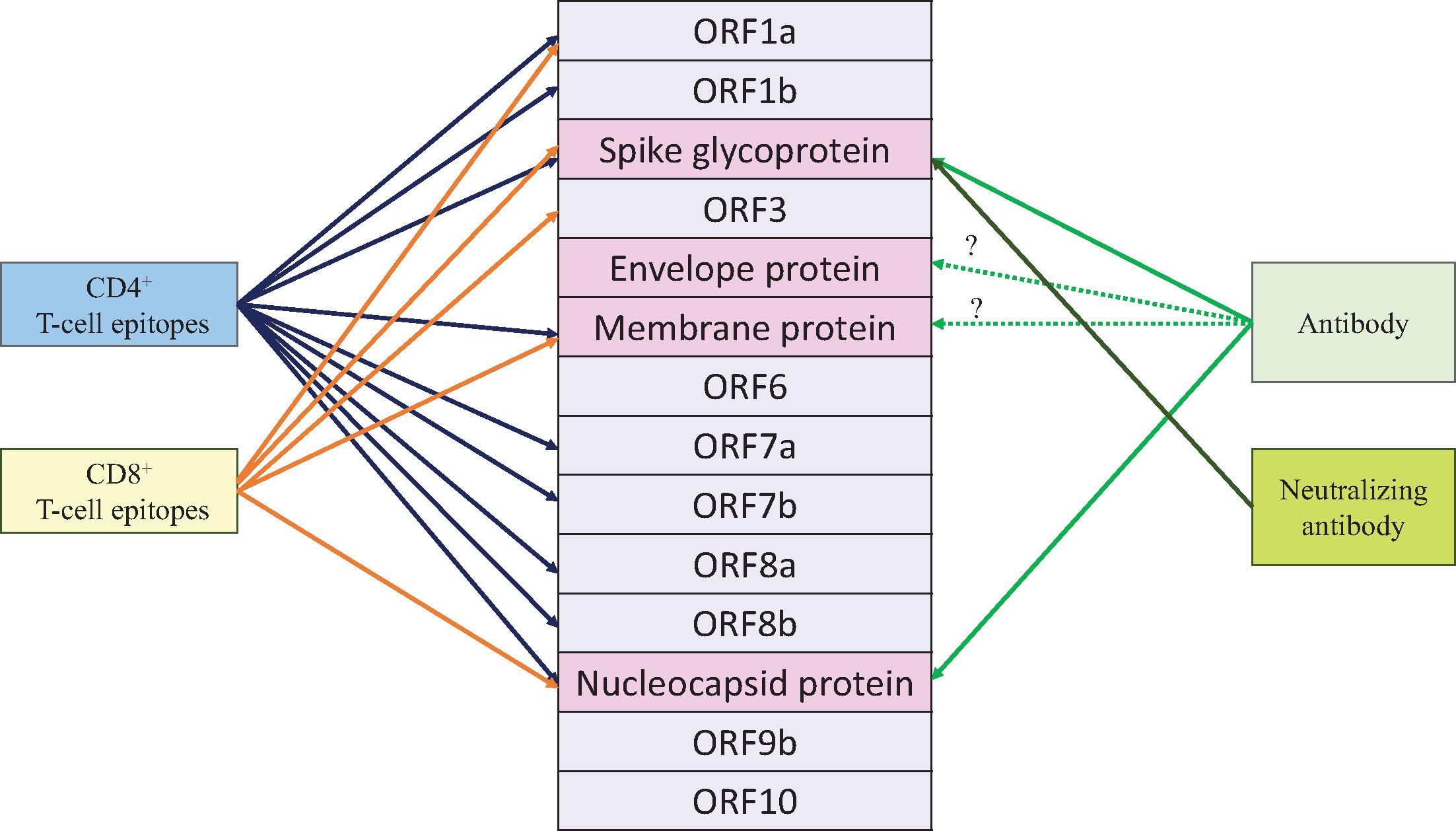
The neutralizing antibodies produced by B lymphocytes and plasma cells play a critical role in preventing SARS-CoV-2 infection in uninfected individuals and restricting viral replication in those already infected. Initial immunoglobulin M (IgM) antibody responses are often of limited binding avidity and functional capacity. The evolution of these responses to include other antibody isotypes with greater avidity and broader functionality (including IgA and IgG subtypes) requires the engagement of T helper (Th) cell lymphocytes. Antigens are presented in the form of short peptides by dendritic cells, macrophages, and B-lymphocytes. Presentation occurs in the context of major histocompatibility complex (MHC) (human leukocyte antigen [HLA]) determinants displayed on the cell surface together with appropriate costimulatory molecules. This process initiates the selection of antigen-specific naïve B and T lymphocytes for further clonal expansion ( Fig. 3.4 ).
Variations in the character and timing of these responses give us clues to the relationship and relative roles of the innate and adaptive immune systems in defenses against COVID-19. Recovery is delayed in the absence of IgG responses (in persons with agammaglobulinemia), with cases reported in which disease progression continued until convalescent serum replacement therapy was administered. Poor outcomes have been reported for individuals requiring treatment with anti–B-cell monoclonal antibodies, with the clinical severity of illness dependent on the extent of the humoral immune deficit. Early decreases in the numbers of lymphocytes or dendritic cells (hindering the clonal expansion of pathogen-specific T and B cells) are similarly associated with sustained viral replication and poor clinical outcomes. , These findings establish a critical role for neutralizing antibodies in defenses against SARS-CoV-2 infection.
Antibody Responses
Two key studies confirm the importance of the SARS-CoV-2 spike protein receptor binding domain (RBD) as a critical antigen for host immune responses. The first study found that RBD antibody accounted for 90% of neutralizing activity in convalescent patient sera. The second found that most convalescent sera with high neutralizing titers specifically targeted the spike protein and its RBD. These findings are consistent with current understanding of the role of the viral spike glycoprotein mediating cell entry by binding to the human angiotensin-converting enzyme-2 (ACE2) receptor. , Additional cross-sectional studies of hospitalized patients acutely infected with COVID-19 detected RBD-specific IgG neutralizing titers within a week of PCR diagnosis, with their magnitude associated with disease severity. In contrast, antibodies against the SARS-CoV-2 nucleocapsid protein are generally nonneutralizing and not associated with severity. These studies confirm the importance of the spike protein as a critical antigen for a beneficial host immune response.
Although SARS-CoV-2 is a new pathogen, it belongs to a yet-evolving extended family of coronaviruses. Researchers have hypothesized that prior immunity to related pathogens might explain the wide diversity of SARS-CoV-2 clinical outcomes. Several studies have examined cross-reactivity of convalescent SARS-CoV-2 patient sera to previously characterized viruses. One study of the spike antigen found no cross-reactivity with the highly homologous pre-pandemic bat coronavirus WIV1-CoV. Another found little correlation between responses to the RBDs of SARS-CoV-2 and the endemic HKU1 and NL63 human coronaviruses or to antigens of influenza or respiratory syncytial virus. A related hypothesis posits that even in the absence of detectable cross-reactivity, individuals with immune memory to related viruses might acquire adaptive responses to SARS-CoV-2 more rapidly. , However, a large study characterizing spike RBD antibody kinetics and isotype profiles in SARS-CoV-2 cases and pre-pandemic controls demonstrated no cross-reactivity with RBDs of Middle East respiratory syndrome coronavirus (MERS-CoV) or endemic human coronaviruses, although cross-reactivity was indeed found for SARS-CoV-1. Together, these findings suggest that common prior viral infections, including endemic human coronaviruses, do not influence the functional evolution of immunity to SARS-CoV-2.
T-Cell Responses and Repertoire
T cells can be characterized according to expressed cell surface proteins detected by flow cytometry. The largest T-cell population, CD4+ helper lymphocytes, recognize peptide fragments from virus proteins complexed with MHC class II molecules ( Fig. 3.5 ). CD4+ T cells can be differentiated further into T follicular helper (Tfh), Th1 or Th2 cells, and others. Tfh cells are essential for antibody affinity maturation and isotype switching. They provide help to B cells in lymph nodes and influence naïve B cells to differentiate to become antibody-producing plasma cells. Tfh cells also are able to provide help to accelerate responses in cases of reinfection or in vaccinated individuals. Th1 cells show antiviral properties such as IFN-γ production. T cells that are already optimized as a result of previous viral antigen exposure and often reside in respiratory epithelium are termed resident memory T cells (Trms). Their cellular properties and anatomic location facilitate rapid responses to infection. The Th2 signature pathway for CD4+ cells has not been a significant feature of SARS-CoV-2 infection. This is fortunate, given its relationship to antibody-dependent disease enhancement and cytokine profiles associated with poor clinical outcomes in other viral diseases, including the experience and lessons learned from respiratory syncytial virus (RSV) infection after formalin-inactivated RSV vaccine administration. , CD4+ T cells additionally recruit innate immune cells to sites of infection and support the clonal expansion of CD8+ T cells. CD8+ T cells in turn can eliminate infected cells through the cytolytic activities of granzyme, perforin, IFN, and TNF-a.
Several elegant studies have examined adaptive immune responses and SARS-CoV-2 epitopes at different points of time in COVID-19 disease, from acute infection to convalescence or terminal illness. Two studies paved the way for vaccine development by demonstrating that SARS-CoV-2 infection induces natural immunity and protects against reinfection in nonhuman primates and that neutralizing antibodies against the SARS-CoV-2 spike protein played a key role in protection. , T-cell responses to the spike protein can be detected within 7 days of infection; these are followed by B-cell responses associated with symptom onset (IgM and IgA by days 5–7, IgG by days 7–10). Antibody and T-cell levels decline after the acute phase of infection; serological memory is maintained by a small number of long-lived bone marrow plasma cells that constitutively secrete antibody and subsequently provide accelerated booster responses after reexposure.
Studies of T cells in patients from the SARS-CoV-1 outbreak in 2003 showed evidence of long-lived recognition of the nucleocapsid (N) structural protein of the virus; when similar assays were performed in convalescent COVID-19 patients, there was evidence of CD4+ and CD8+ T-cell recognition of multiple epitopes on the SARS-CoV-2 N protein as well. In studies with all proteins of SARS-CoV-2 represented, the transmembrane (M) and spike (S) proteins were codominant, with N proteins indicating a different pattern of response to epitopes than with the SARS-CoV-1 outbreak in 2003. A small study of 12 patients characterized cellular immunity, antibody levels, and respiratory SARS-CoV-2 viral load by PCR cycle time and demonstrated that moderate and severe disease status had higher SARS-CoV-2 antibody responses compared with mild clinical syndromes in which early IFN-g production indicated a T-cell response correlated with mild disease and a shorter illness. More recently in a study of nearly 100 convalescent COVID-19 patients evaluating T-cell immunodominance, the S and M SARS-CoV-2 proteins determined CD4+ helper cells’ functional roles for both B-cell help producing RBD antibodies and to CD8+ T-cell responses. CD8+ T cells also have been found to have an important role in decreasing severity of illness; in studies using peptide pools, CD8+ cells are notable for a broad response to viral protein epitopes albeit with different patterns of immunodominance compared with CD4+ cells. CD4+ cell responses to SARS-CoV-2 antigens are most robust in persons recovering from mild COVID-19 disease, whereas antibody production and killing of infected cells by CD8+ T cells predominate in persons with severe disease. Studies with candidate vaccines have demonstrated that SARS-CoV-2 spike induces multiple arms of the immune system, including specific CD4+ and CD8+ T cells and nonneutralizing antibodies that mediate antibody-dependent cytotoxicity. ,
Coverage of Emerging Variants
As SARS-CoV-2 propagates among the global human and nonhuman population, the virus continues to accumulate mutations as a result of natural evolution and immune pressure. Although coronaviruses have been reported to be 10-fold less error-prone than other RNA viruses because of the presence of a proofreading replicase enzyme, natural viral evolution during the pandemic has spawned SARS-CoV-2 variants with distinct mutations in the spike protein. For these emerging variants, we have different levels of scientific understanding of their transmissibility, pathogenicity, and ability for immune escape, though undoubtedly the variants have significantly changed our understanding and concerns regarding the COVID-19 pandemic. Variants were noted, beginning with the notable D614G spike protein mutant that has shown a modest ability for faster spread, and the mink variant with multiple mutations (e.g., “cluster 5”) that demonstrate spillover transmission across species and highlights the risk for incrementally evolved SARS-CoV-2 viruses with broad host range and/or greater pathogenicity. More recently the global public health community has increased the monitoring of emerging SARS-CoV-2 variants more purposefully with stepped up genomic surveillance and a classification scheme of (1) variant being monitored-primarily where data indicates association with increase in transmissibilty or disease severity but surveillance shows very low levels of circulation; (2) variant of interest—primarily with a predicted increase in transmissibility or disease severity; (2) variant of concern—primarily with evidence of an increase in transmissibility, pathogenicity (i.e., causing more severe disease), significant reduction in antibody neutralization, and vaccine effectiveness of treatments or vaccines; and (3) variant of high consequence—with clear evidence that prevention measures or medical countermeasures have significantly reduced effectiveness. , Among the prominent identified variants: alpha (B.1.1.7, first detected in the United Kingdom), beta (B.1.351; first detected in South Africa), and gamma (P.1; first detected in Brazil), were variants of concern but are now being monitored, and current variants of concern include delta (B.1.617.2 and AY lineages; first detected in India), but no identified variants of high consequence. , , The latest nomenclature uses Greek letters for variants of concern and is a departure from the influenza virus naming convention using place of origin and critical mutations.
One study of convalescent patient sera found cross-neutralization of both wild-type and D614G mutant SARS-CoV-2 spike antigens. However, immune evasion was thought partly responsible for a resurgence of COVID-19 cases in Manaus, Brazil in November 2020, which occurred despite previously high levels of infection. The resurgence was temporally associated with the emergence of new SARS-CoV-2 lineage (P.1) with three mutations in the spike protein, potentially increasing ACE2 binding and reducing neutralization by wild-type convalescent sera. Partial antibody resistance also has been described for variants alpha and beta causing second waves of infection in the United Kingdom and South Africa. ,
Interpretation of these findings is handicapped by an incomplete understanding of the relationship of neutralizing antibody titers to protection from infection and/or disease. Neutralization assays may use either authentic SARS-CoV-2 viruses, or pseudoviruses in which specific SARS-CoV-2 proteins are expressed in other viral vectors such as human immunodeficiency virus-1 (HIV-1) or vesicular stomatitis virus, using well-characterized assay systems. A prototypical study early in the pandemic that collected convalescent sera from 149 patients of varying severity found a wide range of half-maximal neutralizing titers (NT 50 s). Only 1% of samples showed very high titers (>5000); the majority were less than 1000, and one-third were less than 50. Interestingly, nonhuman primate challenge studies have reported circulating neutralizing antibody titers of 1:20 or greater in animals protected from SARS-CoV-2 infection. Generally, however, the levels in most convalescent sera are broadly comparable to those after full vaccination in individuals receiving mRNA, adenoviral vector, or whole-virus inactivated vaccines.
To date, all variants tested have been neutralized in vitro by immune sera from individuals vaccinated with mRNA, viral vector, or inactivated vaccines, although reduction in neutralization titers have been observed with particular variants. , Studies have shown that certain mutations and variants (E484K mutation in particular) are able to escape neutralization by convalescent plasma and monoclonal antibodies that target single epitopes. , , Vaccine-elicited sera were able to neutralize engineered SARS-CoV-2 viruses containing key variant spike mutations in vitro, such as E484K, L452R, and N501Y mutations. , Studies of cellular immune responses to SARS-CoV-2 variants in recovered COVID-19 patients demonstrated that CD4+ and CD8+ T cells have broad responses to multiple epitopes compared with neutralizing antibodies to the S protein involved with humoral immunity; this could bode well for the durability of vaccine-induced T-cell responses, given ongoing mutations of SARS-CoV-2 virus that have demonstrable effects in vitro on the amount of antibody needed to neutralize SARS-COV-2.
Reduced efficacy against clinical disease was observed in late-stage vaccine clinical trials in regions with high circulation of emerging variants. Interim analysis in January 2021 for an adenoviral vector vaccine (Ad26.COV2.S) in a single-dose regimen reported 66% efficacy overall against moderate to severe COVID-19, with 72% efficacy in the United States and 66% efficacy in Latin America cohorts, but efficacy dropped to 57% in the South African cohort, wherein 95% of the accrued cases were caused by the beta variant. , Similarly, phase III results for the adjuvanted protein subunit vaccine NVX-CoV2372 showed 89.3% efficacy in the UK cohort, in which more than 50% of accrued cases were caused by the alpha variant; efficacy in the South African cohort was 51%, with 93% of the accrued cases caused by the beta variant. On the other hand, reassuring real-world vaccine effectiveness data have been accumulating from national immunization programs for COVID-19 vaccines. Two doses of BNT162b2 mRNA vaccine demonstrated high real-world vaccine effectiveness against circulating variants of concern: in Israel, BNT162b2 demonstrated 90% to 96% effectiveness against SARS-CoV-2 infection, asymptomatic infection, symptomatic COVID-19, severe and critical hospitalizations, and deaths during the period of high circulation (94%) of the alpha variant. Similarly, from Qatar’s national immunization program, a two-dose regimen of BNT162b2 demonstrated 89% effectiveness against alpha variant infection; 75% against beta variant infection; and 100% against severe, critical, or fatal COVID-19 caused by alpha or beta variants. A two-dose regimen of mRNA-1273 demonstrated 100% effectiveness against alpha infection; 96% against beta infection; and 96% against severe, critical, or fatal COVID-19 caused by these two variants. In the UK national immunization program, two-dose BNT162b2 mRNA effectiveness was 93.4%, with alpha cases and 87.9% with delta cases, and two-dose ChAdOx1 effectiveness was 66% with alpha cases and 59.8% with delta cases. Vaccine effectiveness against the delta variant has been reported from national immunization programs in the United Kingdom and Canada. A two-dose regimen of BNT162b2 or ChAdOx1 vaccine demonstrated between 83% and 88% and 61% and 67% effectiveness, respectively, against symptomatic COVID-19. Scotland reported 79% and 60% effectiveness against SARs-CoV-2 infection; Canada reported that a two-dose regimen of BNT162b2 was 95% effective against hospitalization or death. Data for two doses of an inactivated SARS-CoV-2 vaccine (Coronavac) from Chile indicated 66% to 90% effectiveness against COVID-19, hospitalization, intensive care unit (ICU) admission, and COVID-19–related death. Brazil reported that in adults older than 70 years, effectiveness was 42% against symptomatic COVID-19, 59% against hospitalization, and 71% against death. Systematic reviews of vaccine effectiveness data across multiple vaccine platforms, dosing intervals, varied geographies and assorted SARS-CoV-2 variants in circulation are now available. , In addition, the first real world data are now available for effectiveness of maternal vaccination using mRNA vaccines: 2-dose COVID-19 mRNA maternal vaccination during pregnancy showed 61% VE against COVID-19 hospitalization in infants <6 months, during a period of delta and omicron variant predominance in the USA. Although no substantial clinical evidence of variant escape from vaccine-mediated protection has emerged, vaccine manufacturers and regulatory bodies are building clinical evidence and frameworks for redesigning vaccines and/or booster vaccines using either the ancestral strain of SARS-CoV-2 or the emerging variants, if the need arises. The most recently emerged SARS-CoV-2 variant – oicron (B.1.1.529 and BA lineages) has at least 30 mutations in the spike protein, half of which are in receptor binding domain , (RBD) that interacts with the host ACE2. receptor Furthermore, omicron does not appear to be the result of a linear progression from its immediate predecessor – delta, prevalent in 2021 – in an easily predictable fashion, but rather it appears to have evolved from the alpha variant prevalent in 2020. In part due to it’s significantly divergent sequence that enables immune evasion, omicron is highly transmissible and has spread rapidly to become the dominant circulating strain in most parts of the world. , Initial observations indicate a comparatively mild clinical phenotype, and multiple studies have shown that neutralizing antibody responses are substantially diminished post-2-dose vaccination while T-cell responses appear to be preserved, and a 3rd vaccine dose confers protection against omicron-related hospitalization and severe disease. , ,
Durability of Immunity
Early evidence indicated that protective immunity against seasonal CoVs is short term, that is, lasting 6 to 12 months, based on data derived from a 35-years-long epidemiological study examining frequency of reinfection with seasonal CoVs—NL63, 229E, OC43, and HKU1 in a cohort of individuals—as a measure of natural immunity. In a 2020 large US serosurvey of more than 30,000 individuals who tested positive for SARS-CoV-2, more than 90% had detectable neutralizing antibody responses and antibody titers were stable over 3 months with modest declines at 5 months. Another study measured risk of reinfection in a cohort of 12,000 health care workers based on the presence of antibodies to SARS-CoV-2 antibodies and found that postinfection immunity is associated with protection from reinfection for most individuals, for at least 6 months. In population-level serosurveillance in Iceland of PCR-positive persons (n = 30,576), more than 90% remained seropositive 120 days after diagnosis, with no decline detected in antibody levels. Two recent studies with convalescent patients showed that antibodies to SARS-CoV-2 persist for nearly 1 year after infection. One study with 77 convalescent individuals monitored regularly over 1 year showed that SARS-CoV-2 spike-specific antibodies were detectable 11 months after infection, showing a rapid decline for 4 months and a slower decline in the subsequent 7 months; furthermore, spike-specific bone marrow plasma cells persisted for 7 to 8 months in a subset of this convalescent cohort, indicating the stable presence of memory B cells. A similar second study in convalescent patients (n = 63), approximately 40% of whom were vaccinated with at least one dose of mRNA vaccine, also demonstrated the stable presence of SARS-CoV-2 spike-specific antibodies and memory B cells 6 to 12 months after infection. Very few reinfections with the wild-type or ancestral SARS-CoV-2 strain have been reported, and generally the subsequent disease episode has been less severe than the first, suggesting benefit from the protective immune responses triggered during the first infection.
For SARS-CoV-2 mRNA vaccines authorized for emergency use, data on longer-term persistence of vaccine-elicited immune responses will become available as the participants from the phase III vaccine clinical studies are followed for 2 years after primary vaccination. An interim readout of immune persistence has been reported for mRNA-1273 and BNT162b2 mRNA vaccines, suggesting that antibody binding and neutralization titers were maintained for 3-6 months after primary vaccination among study participants. An Ad vector vaccine against a related coronavirus, ChAdOx1-MERS, showed that vaccine-elicited neutralizing antibodies waned from peak postprimary immunization levels and remained stable above baseline for 1 year or longer. Since COVID-19 mass vaccination programs began in late 2020, real world evidence for the duration of protection has been accumulating. The current state of understanding can be represented by the data available for vaccine effectiveness (VE) of the mRNA vaccine BNT162b2 against the latest circulating variant – omicron: against omicron infection – 2-doses of BNT162b2 conferred limited protection (~50% VE) for only one month and 3-doses increased VE up to 70-80%, with waning observed after the first few months; against omicron-mediated symptomatic COVID-19 – 2-doses of BNT162b2 demonstrated ~40-70% VE and 3-doses increased VE up to ~55-85%, both waning after the first few months; and finally against omicron-mediated hospitalization – 2-doses of BNT162b2 showed ~55-80% VE before 6 months and ~35-80% VE after 6 months after vaccination; 3-doses increased VE to ~75-90% with waning after the first few months. , , ,
Correlates of Protection
Correlates of protection have been established for many different viral infections, such as influenza, measles, and hepatitis A and B viruses, and bacterial infections, such as pneumococcal and meningococcal disease, and are typically based on the level of antibody that is acquired from vaccination or natural infection that is able to significantly reduce the risk for infection or reinfection. , However, correlates of protection have yet to be defined for COVID-19, , in part because antibody and cellular immune responses may differentially affect risk for infection versus progression to severe disease. For a novel disease such as COVID-19, systems serological studies might potentially yield novel immune correlates based on composite antibody and cellular immune measurements. However, more familiar immune correlates, such as spike-specific neutralizing antibody titers, are being deduced from the immunogenicity data reported from the handful of COVID-19 vaccines authorized for emergency use during the pandemic. Modeling the relationship between vaccine-elicited neutralizing antibody titers and observed protection against SARS-CoV-2 infection, normalized against convalescent serum standards, , at least one study has deduced that a 50% protective titer was 20% of the convalescent level; 50% protective titer against severe disease was even lower, at 3% of the convalescent titer. mRNA vaccines ranked highest in terms of protective efficacy, followed by a protein vaccine; adenoviral vector vaccines ranked at or below convalescent protective titers, and inactivated vaccines ranked the lowest. Finally, modeling the duration of protection predicted that although protection against severe disease will persist, loss of protection against SARS-CoV-2 infection is likely over the first 250 days modeled; in addition, the model predicts erosion in protection against SARS-CoV-2 variants that elicit reduced neutralization titers. Taken together, this would suggest the need for booster vaccine doses potentially on an annual basis to build up population protection against SARS-CoV-2. Development of an immune correlate of protection will catalyze rapid vaccine development for additional populations such as children younger than 12 years, pregnant and lactating women, and those with immunosuppressive conditions, in whom traditional field efficacy trials would be unfeasible. Potentially, it would also streamline the development of vaccines with updated compositions, which might be necessary to combat novel SARS-CoV-2 variants.
Systems Serology and Systems Immunology
Systems serology aims to define the breadth and depth of humoral responses to vaccine antigen(s) by measuring both polyclonal antibody features (antigen-binding portion, Fab and functional properties, and Fc portion) to clarify immune mechanisms and define multifactorial correlates of protection that can help assess vaccine candidates. Molecular techniques have enabled higher resolution understanding of response determinants on both sides of the antigen–antibody reaction. Mining the antigenic proteome with antigen arrays enables screening for unique target epitopes—both linear sequences and conformationally defined epitopes, and the influence of posttranslational modifications such as glycosylation—that may have differential roles across the disease course and reveal the complex nature of elicited antibody response. On the other side, parallelized investigations of antibody downstream signaling, for example, activation of natural killer (NK) cells, monocytes, macrophages, neutrophils, dendritic cells, and effector responses, such as degranulation, cytokines secretion, cytotoxicity, and phagocytosis, help elucidate the complex information relay that triggers the comprehensive immune response to an antigen or pathogen and the dysfunction that often characterizes the deleterious inflammatory response. For a brand-new pathogen such as SARS-CoV-2, this could help identify biomarkers for predicting the COVID-19 trajectory in infected persons. A comprehensive genome map of SARS-CoV-2 created by consensus of experts may facilitate such studies.
Four distinctive immune response profiles that predicted divergent courses of COVID-19 within 10 days of SARS-CoV-2 infection were deduced from longitudinal immune profiling of peripheral blood mononuclear cells and plasma samples from 113 patients with moderate (n = 80) or severe (n = 33) COVID-19. An aberrant immune response with early, elevated proinflammatory cytokines was seen in patients with severe COVID-19 and poor clinical outcomes. In all COVID-19 patients, an overall increase in innate cell lineages such as monocytes, low-density neutrophils, and eosinophils was seen, with a parallel decrease in CD4+ and CD8+ T-cell levels. Similarly, the ratio of neutrophils to lymphocytes emerged as a prognostic biomarker of COVID-19 severity and organ failure from comprehensive immunophenotyping of peripheral blood in 42 convalescent patients with divergent clinical trajectories of SARS-CoV-2 infection and COVID-19 (moderate n = 7, severe n = 28, recovered n = 7) that showed perturbations in multiple leukocyte populations, which distinguished cases of severe COVID-19 from healthy donors, cases of moderate COVID-19, and patients who have recovered from COVID-19.
A study by Mathew et al. found three distinct immunotypes in hospitalized COVID-19 patients that represent patterns of different suboptimal responses to infection: (1) robust activation of CD4+ T cells and highly activated or exhausted CD8+ T cells, (2) less robust CD4+ T cells and highly functional effector-like CD8+ T-cell responses, and (3) a lack of lymphocyte response.
In a cohort of 22 SARS-CoV-2–positive patients (recovered, n = 12; deceased, n = 10), Ateyo et al. identified distinct humoral profiles comprising five antibody features that could differentiate between patients who went on to recover from or succumb to COVID-19. Although the two patient classes showed equivalent magnitude of the responses, convalescent patients showed a more spike-focused humoral response, including phagocytic and complement activity versus deceased patients who showed stronger nucleocapsid (N)-specific responses, poorly coordinated RBD-specific antibody-dependent complement deposition and NK cell functions. A prototypical biomarker—spike-to-N ratio—of humoral response was confirmed in a larger validation cohort of acutely infected individuals (recovered, n = 20; died, n = 20). Similarly, neutralization potency indices predicted disease severity and survival in a cohort of 113 patients infected with SARS-CoV-2 stratified by disease severity and outcomes (i.e., nonhospitalized, hospitalized, intubated, immunosuppressed, and deceased), anti-RBD antibody levels and neutralization titers, indicating that potent SARS-CoV-2–specific neutralizing antibodies appear to increase survival and may protect against reinfection with variants of SARS-CoV-2.
Finally, Dan et al. combined five immune components to devise a composite measurement of SARS-CoV-2 immune memory: RBD-specific IgG, IgA, memory B cells, and SARS-CoV-2–specific CD4+ and CD8+ T cells. They measured circulating SARS-CoV-2 antigen-specific antibodies, memory B cells, and CD8+ and CD4+ T cells for more than 6 months after infection in a cohort of patients with COVID-19 (n = 188) across a range of disease, including asymptomatic, mild (nonhospitalized), moderate (hospitalized), and severe (hospitalized). They determined that at 1 to 2 months after infection, 59% of individuals were positive for 5 of 5 components, and at 5 months or longer after infection, 40% were positive for 5 of 5 components; however, 96% of individuals were positive for 3 of 5 components.
Immune Dysregulation and Immunopathology
A significant minority of adult COVID-19 patients develop hypoxemic respiratory failure, with diffuse alveolar damage, inflammatory infiltrates, and intravascular thrombosis (involving both large and small blood vessels), often associated with lymphopenia and markedly increased levels of inflammatory markers, including C-reactive protein, d -dimers, interleukin-1 (IL-1), and IL-6. , , Similar clinical and immunological features have been reported in other severe viral pneumonias, including pandemic influenza, severe acute respiratory syndrome (SARS), and Middle East respiratory syndrome (MERS).
In a study of adaptive immune responses with serial paired sampling of systemic and lung compartments in severely ill COVID-19 patients compared with uninfected controls, investigators noted a chemokine signature CCL2-CCR2 associated with myeloid cell recruitment; cells of the innate immune system that in this study were associated with older patients and higher mortality whereas patients with Trm signature T-cell profiles were associated with younger age and lower mortality. Replication of the study with larger numbers will be needed to differentiate among the effects of age, cell profiles, and mortality.
A study of coendemic diseases affecting T-cell function and SARS-CoV-2 described clinical features of 95 hospitalized COVID-19 patients of whom 38 patients were infected with HIV, tuberculosis (TB), or both and compared the CD4+ T-cell responses to 38 hospitalized patients without SARS-COV-2 infection. Severe COVID-19 disease was associated with altered cellular immunity with deficits of CD4+ cell function potential and a reduced capacity to proliferate.
Antibodies may also participate in disease exacerbation, particularly through interactions with cells expressing Fc receptors. These risks, described as antibody-dependent enhancement (ADE) of disease, have been observed with SARS-CoV-1. It is important to understand the balance between Fc signaling that promotes protective immunity and that which promotes inflammatory pathological processes and whether systems serology can identify those at risk. To date, there has been no evidence detected of ADE resulting from SARS-CoV-2 vaccines in preclinical studies or nonhuman primate studies. Similarly, no evidence of ADE emerged in a phase III clinical study of an mRNA vaccine against SARS-CoV-2 after a median 2 months of follow-up. Instead, it appears that immunopathological processes in severe COVID-19 mainly occur as a result of aberrant or excessive innate responses.
Clinical trials of antiinflammatory therapies in patients with severe COVID-19 can provide insights into the underlying pathological mechanisms.
Corticosteroids
As part of the RECOVERY trial, 2104 hospitalized COVID-19 patients assigned to receive dexamethasone 6 mg daily were compared with 4321 receiving usual care. Findings differed substantially according to the degree of illness severity. The greatest benefit was observed in patients receiving invasive mechanical ventilation at the time of randomization, in whom adjunctive dexamethasone reduced the risk for death from 41% to 29% (RR, 0.64; 95% CI, 0.51–0.81). A smaller but still significant benefit was found in patients receiving oxygen without invasive mechanical ventilation (26% mortality in controls vs. 23% in dexamethasone recipients; RR, 0.82; 95% CI, 0.72–0.94). No benefit was found for patients not receiving respiratory support at randomization; indeed, in this subgroup, the trend was toward increased mortality risk attributable to dexamethasone (RR, 1.19; 95% CI, 0.92–1.55). Thus corticosteroids and IFN are each of greatest benefit to patients at the opposite polar extremes of disease stage and severity.
Corticosteroids exert dose-dependent antiinflammatory and immunosuppressive effects on nearly all immune cells, reducing production of proinflammatory cytokines, inhibiting cellular microbicidal responses, and increasing the risk for many bacterial, fungal, and viral infections after prolonged use. Nonetheless, corticosteroids have become essential adjunctive treatments for several serious or life-threatening infections. In Pneumocystis jiroveci pneumonia, early adjunctive steroid treatment improves survival and decreases the need for mechanical ventilation, particularly in patients with the most profound immune dysregulation, for example, acquired immunodeficiency syndrome (AIDS) patients not yet on antiretroviral therapy (ART). In patients beginning treatments for both AIDS and TB, corticosteroids reduce the risk for immune reconstitution inflammatory syndrome (IRIS), or alternatively can be used to treat IRIS once it has occurred. , IRIS is characterized by clinical worsening despite microbiological improvement; it is often accompanied by extrathoracic suppurative lymphadenopathy and is attributed to ART-associated recovery of immune function. Finally, a systematic review in 1997 and a formal meta-analysis in 2013 concluded that corticosteroids conferred a survival advantage in non-HIV central nervous system and pericardial TB and that in pulmonary TB, steroids hastened the resolution of constitutional, radiographic, and pulmonary function abnormalities. , The findings are striking in that, like in severe COVID-19, adjunctive corticosteroids provide a clinical benefit in selected patients despite their detrimental overall effects on host defenses against these infections.
Interleukin-6 Receptor Blockade
Interest in the role of IL-6 in COVID-19 pathogenesis in large part reflects the successes of IL-6 receptor blockade in the hyperinflammatory complications such as cytokine-release syndrome and macrophage activation syndrome associated with chimeric antigen receptor (CAR)-T cell therapy. Although serum levels of IL-6 are indeed elevated in severe COVID-19, they are not strikingly so. A systematic review published in late 2020 compared serum IL-6 concentrations in 1245 patients with severe or critical COVID-19 to 8159 others with life-threatening conditions. The mean IL-6 level in COVID-19 patients was 36.7 pg/mL (95% CI, 21.6–62.3 pg/mL). Mean concentrations were nearly 100 times higher in patients with CAR-T cytokine release syndrome, 27 times higher in patients with sepsis, and 12 times higher in patients with acute respiratory distress syndrome unrelated to COVID-19. One early study of IL-6 receptor blockade in hospitalized COVID-19 patients (COVACTA) found no benefit. However, two subsequent studies—RECOVERY and REMAP-CAP—found that IL-6 receptor blockade improved clinical outcomes, including risk for progression to invasive mechanical ventilation and death. , The contrary findings of these studies may be due to the use of corticosteroids, which were given to a greater proportion of patients in the later studies. It appears the two treatments target complementary pathways and that the modest doses of dexamethasone given in COVID-19 are insufficient to fully inhibit the effects of IL-6.
Signaling Pathway Inhibition
Successful viral pathogens face a challenge to take command of multiple cellular biosynthetic and metabolic processes using only limited genomic space. A strategy common to several viruses, including SARS, SARS-CoV-2, MERS, and pandemic influenza, is to seize control of two interrelated pathways—(Janus kinase–signal transducer and activator of transcription (JAK-STAT) and mammalian target of rapamycin (mTOR)—to promote cell activation, division, growth, and survival. Pharmacological inhibitors of these pathways therefore have two potential therapeutic properties, inhibiting viral replication and reducing immunopathological processes. In one study in 1033 hospitalized COVID-19 patients, baricitinib, a Jak1/2 inhibitor, shortened recovery time and tended to improve survival. Benefit was greatest in the most seriously ill subset of patients. A second study in 83 seriously ill patients found baricitinib improved survival and inhibited viral replication. A similar study in 289 hospitalized patients found that treatment with the Jak inhibitor tofacitinib led to a lower risk for death or respiratory failure through day 28 than placebo. Antiinflammatory and antiviral effects of mTOR inhibitors have been reported in influenza, but studies in COVID-19 have not yet been reported.
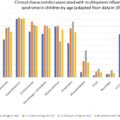
Pathogenesis of Multisystem Inflammatory Syndrome in Children
Children infected with SARS-CoV-2 are at reduced risk for pneumonia compared with adults but are at increased risk for subsequent multisystem inflammatory syndrome (MIS-C). This syndrome, which resembles Kawasaki disease, is a late phenomenon driven by inflammatory processes peaking at a time thought initially to be well after SARS-CoV-2 levels have declined. Its manifestations are mainly extrapulmonary, with vascular damage associated with autoantibodies directed against a specific set of autoantigens. , The antiinflammatory immunotherapy that is effective in Kawasaki disease appears similarly effective in MIS-C. The approach combines high-dose intravenous immunoglobulin that activates inhibitory Fc-receptors with administration of high-dose corticosteroids. In some cases, an anti–IL-6 receptor antibody or recombinant IL-1–receptor antagonist is also used. The report of a single patient treated with larazotide, a zonulin inhibitor, followed studies of a cohort of children with prolonged residence of SARS-CoV-2 in the gastrointestinal tract, with gut permeability changes and SARS-CoV-2 antigen presence in plasma, that provide further insight into the long-term manifestations in children.
The Global Pandemic and Questions for Future Research
Advanced age is a consistent risk factor for severe disease and death and may indicate that the cellular immune system and immunosenescence are in play and important considerations for early prediction of poor outcomes and for constructing highly effective and durable vaccines. , Our ability to predict the risk for progression to severe or life-threatening COVID-19 in individual patients presently is limited. Although the hypothesis that severe disease is due to delayed IFN responses, heightened early viral replication, and exaggerated subsequent late innate responses is plausible, it remains to be confirmed and its underlying mechanisms determined. Further refinements will undoubtedly occur, such as a report documenting that some of the risk for severe disease in men and in the elderly can be explained by the increased occurrence of anti-IFN autoantibodies.
The burden of coendemic diseases, such as Mycobacterium tuberculosis (Mtb) infection, that have latent phases that may progress to clinical disease and are prevalent in low-income countries may also change. For example, a study of coendemic diseases affecting T-cell function and SARS-CoV-2, described clinical features of 95 hospitalized COVID-19 patients in which 38 patients were infected with HIV, TB, or both and compared the CD4+ T-cell responses to 38 hospitalized patients without SARS-COV-2 infection. Severe COVID-19 disease was associated with deficits of CD4+ cell functional potential and a reduced capacity to proliferate. Larger studies of CD4+ cell function will be needed as the SARS-CoV-2 pandemic increasingly spreads to overlap endemic geographies with HIV and TB coinfections particularly since the investigators described a reduced frequency of Mtb-specific CD4+ cells that may indicate T-cell exhaustion with the potential implications for reactivation of latent TB in a large number of people. However, it was somewhat reassuring that the small number of patients with good control of HIV compared with HIV-negative patients did not have alterations in the functional profile of SARS-CoV-2 CD4+ T cells. Further studies in coendemic geographies are urgently needed to explore and expand on these observations.
The precise timing of interventions is crucial and must take into consideration comorbidities and coendemic diseases. Reliable biomarkers indicating the stage of disease would be extremely helpful. Because the early phase of infection is often asymptomatic, many of the direct antiviral interventions may come too late. Endotypes, defined as distinct molecular profiles based on metabolism, epigenetics, transcription, or immune function, have been proposed to guide the application of personalized immunotherapies. One study, for example, found low serum levels of sphingosine‐1‐phosphate (S1P) to be a strong predictor of ICU admission and mortality in COVID-19. S1P regulates endothelial integrity through its binding to high-affinity G protein–coupled receptors. In models of influenza and SARS-CoV-1 infection, agonists and antagonists of the S1P1 receptor had strong effects on cytokine storm and lethality. , Our understanding of specific endotypes in COVID-19 is incomplete. Interventions with greater specificity may be possible, for example, preserving the beneficial effects of cytokine families with pleiotropic functions such as IFNs and IL-6 while minimizing their proinflammatory signals. This may be achieved by employing for therapy specifically those cytokine family members with the lowest inflammatory potential (e.g., IFN-III rather than type I) or by targeting cytokine signaling modalities that specifically promote inflammation. Similarly, pharmacological blockade of some but not all signaling pathways downstream of key cytokines may represent interventions with more specific effects.
Monitoring of vaccine efficacy and routine sequencing of SARS-CoV-2 will be essential to guide changes in vaccine constructs over time as expected changes in B-cell epitopes occur or if vaccinated patients develop severe diseases. This will be particularly urgent because it relates to the campaign for global vaccination in increasingly heterogenous populations with differences in comorbidities and coendemic diseases.
Conclusion
The SARS-CoV-2 genome was sequenced and placed in the GISAID repository before the awareness that a global pandemic causing disease (COVID-19), on the scale of the H1N1 influenza pandemic of 1918, was about to occur. In the early days, investigators naturally tried to understand the evolving pathogenesis and clinical profile of the disease by making comparisons to previous coronavirus infections. Human coronaviruses infect toddlers universally in the first years of life, and reinfection is the norm. More lethal coronavirus infections such as MERS and SARS-CoV-1 manifested as lethal full-blown infections with high mortality rates. As of this writing, vaccines created from mRNA, engineered adenovirus constructs expressing the spike protein, and more traditional subunit and inactivated vaccines have demonstrated a range of protective immunity against severe disease, death, and a decrease in spread from person to person. The interplay of innate, humoral, and cellular immunity is now better understood as investigators create models of infection and disease to find better means of protecting people and treating patients exposed to SARS-CoV-2. This chapter gave an overview of the current understanding of the immunology and pathogenesis of SARS-CoV-2 and focused on unique aspects that may provide rich areas of scientific exploration to advance our understanding of this pandemic. The rapid expansion of information on SARS-CoV-2 and COVID-19 clinical disease presents challenges to the student beginning study of this pandemic. We have aimed to provide a framework for understanding the immune system as it relates to a new virus spreading throughout humanity and the current understanding of how the virus interacts with different human hosts.
Acknowledgments
Thank you to Ms. Vaidehi Wadhwa M. Pharm. , Pfizer India Inc.for graphics and editorial assistance . Thank you to peer reviewers: Hernan Valdez, MD, Samuel H. Zwillich, MD, from Pfizer Global Development and Daniel Griffin, MD, PhD, Dickson D. Despommier PhD from Parasites without Borders.
REFERENCES
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree