(1)
Medical Sciences Division Northern Ontario School of Medicine West Campus, Lakehead University, Thunder Bay, Ontario, Canada
Key Topics
Molecular pathology of head and neck cancer (HNC)
Circulating cell-free nucleic acids as HNC biomarkers
Circulating HNC miRNA biomarkers
Circulating HNC cells
Circulating nasopharyngeal carcinoma biomarkers
Key Points
The majority of HNCs are squamous cell carcinomas (HNSCCs) and nasopharyngeal carcinomas (NPCs). The etiologic agents of HNSCC are toxins (tobacco and alcohol) and infectious agents from human papillomavirus (HPV), while those of NPC are mostly Epstein–Barr viral (EBV) infections. HNCs have distinct geographic distribution and molecular pathology.
Circulating biomarkers, especially HNC cells, are potentially valuable in HNSCC staging, prognosis, and prediction of treatment response. While the incidence of HPV-positive HNSCC is on the rise, there is no established diagnostic biomarker for this subtype. There is a need to develop noninvasive biomarkers for accurate detection and management of HPV-positive HNSCC.
Because of the established etiology, there are numerous biomarkers for NPCs. Validated and in clinical practice are serologic and molecular assays targeting EBV biomolecules. Other potentially useful circulating biomarkers of NPCs are the numerous EBV-encoded miRNAs in circulating exosomes.
2.1 Introduction
Head and neck cancer (HNC) is the sixth most common cancer with annual global incidence and mortality being 600,000 and 300,000 cases, respectively. The 2016 estimated incidence and mortality cases for the US are 48,330 and 9,570, respectively. They are more common in males than females, with a male to female ratio of about 3:1. High incidence rate of up to 20 per 100,000 is observed in Central and Eastern Europe, Germany, Denmark, Scotland, Italy, Spain, France, Brazil, Hong Kong, the Indian subcontinent, South Africa, and Australia.
Head and neck cancer comprises an anatomic conglomerate of malignancies that arise from the epithelial lining of the upper aerodigestive tract (excluding the thyroid and parathyroid glands). Collectively, tumors of the oral cavity, nasopharynx, oropharynx, hypopharynx, larynx, nasal cavities, paranasal sinuses, and salivary glands constitute HNC. However, the commonest sites are the oral cavity, where many cases (~250,000) are diagnosed annually, and the oropharynx. It should be noted, however, that the etiologic and molecular features differ considerably among these cancer subtypes. For example, salivary gland tumors are biologically distinct. The majority of HNCs (>90 %) is of squamous cell histology, and hence, many studies refer to them as head and neck squamous cell carcinomas (HNSCCs).
Environmental factors and lifestyle exposures such as tobacco and excess alcohol use are known risk factors of oral squamous cell carcinoma (OSCC). Tobacco use is associated with oral and laryngeal cancers, while alcohol use causes more pharyngeal and laryngeal tumors. Additionally, HPV infection of the oropharynx is an etiologic agent for oropharyngeal squamous cell carcinoma (OPSCC), while nasopharyngeal EBV infections account for the majority of nasopharyngeal cancers (NPC), especially in endemic areas. With increasing education and adherence to cessation of tobacco use, the incidence of non-HPV cancers is declining; however, HPV-mediated OPSCC is on the rise, especially in the Western world.
Modern salvage or organ-sparing surgeries coupled with chemoradiation are improving the quality of life of patients with HNC, although no change in the overall survival rate is accomplished due probably to the concept of field cancerization (i.e., locoregional recurrences are common). Additionally, the 5-year survival rate is ~60 %, and this declines with advancing tumor stage. Thus, biomarkers that can be used for early detection (when treatment including administration of chemopreventive remedies is optimal), prognosis, and therapy selection can improve patient management and hence improve survival rates. Noninvasive screening assays targeting biomarkers in saliva or blood in high-risk groups such as smokers, excessive alcohol users, and those involved in oral sex as well as those in endemic areas of EBV infections could lead to early detection, with possible curative interventions, given that only 33 % of all cases are currently detected at an early stage.
2.2 Screening Recommendations for HNC
There are currently no recommended screening guidelines for HNC, as has been established for breast, colon, and prostate cancers. This situation is partly because of lack of evidence that any screening method increases survival rates. There are also no validated blood- or saliva-based tests to detect early HNCs. There is thus the need to develop validated and cost-effective noninvasive tests for HNC.
Individual health centers have their own protocols as to how to screen the population for early cancer detection. For example, the Memorial Sloan Kettering Cancer Center offers free annual head and neck screening to community members. The authorities here also recommend that patients receive annual head and neck examination from their primary care physicians and dentists. This screening should include at least head and neck examination and inspection of the oral cavity and oropharynx. For high-risk individuals, especially those cured of HNSCC, the National Comprehensive Cancer Network has follow-up guidelines to monitor for possible recurrence or development of second primary tumors. These recommendations are periodic physical examination at the following defined frequencies: every 1–3 months for the first year, every 2–4 months for the second year, every 4–6 months during the third to fifth years, and every 6–12 months thereafter.
2.3 The Need for Noninvasive Screening Tests for HNC
The pathology of premalignant or precursor lesions of HNC is well studied. Mostly, OSCC originates from precursor lesions. Oral leukoplakia, which is a white mucosal lesion in the oral cavity (homogenous), and its variant, erythroleukoplakia that is similar to white lesions mixed with red plaques (nonhomogenous), are premalignant lesions of OSCC. These lesions are associated with risk factors such as tobacco and areca nut use. The prevalence of these lesions varies from 0.1 to 0.5 %, and the rate of progression to oral cancer is ~1–2 % per year. Multiple factors including female gender, size of lesion, anatomic location, and presence of dysplasia or erythroleukoplakia determine lesion progression to malignancy. Additionally, the presence of cancer-associated genetic alterations predicts progression. The risk of these lesions developing into cancer includes mutations in TP53, loss of chromosome 9p, and decreased cytokeratin 4 and cornulin expression, among other factors. Chromosomal loses at 9p (CDKN2A and PTPRD loci), 3p (FHIT and RASSF1A loci), and 17p (TP53 locus) increase the risk of progression. Molecular assays based on salivary washes or buccal swabs for examination of these genetic changes are commendable and will be easily accepted. Such screening assays are also useful because chemoprevention can be offered to delay or reverse disease progression if detected early.
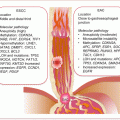
As the original cancer studied by Slaughter and other workers, the molecular pathway or basis of field cancerization in HNSCC is fairly well characterized. The patch-field squamous cell carcinoma pathway of field cancerization has also been well documented. Mutations or other genetic changes in mucosal epithelium create multiple clonal patches. Gain in growth advantage or evasion of growth control, senescence, and apoptosis leads to expansion of patches. Additional mutations or other genetic and epigenetic events in subclones from these clonal patches can lead to the development of invasive cancer. The genetic sequence of events includes (Fig. 2.1):
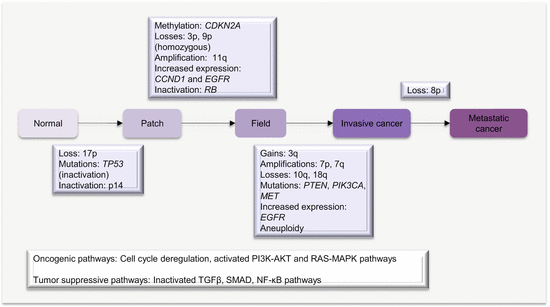
An early loss of chromosome 17p or mutations in TP53 characterizes clonal patches.
The pathway to invasive cancer is further driven by homozygous loss of chromosome 9p, 18q, amplification of 11q, mutations in CDKN2A, and increased expression of CCND1.
Further amplifications of chromosomes 7p, 7q (high-level amplification), and 3q in association with loss of 10q, activate the EGFR, RAS, and/or PI3K/AKT signaling pathways to drive full malignant transformation.

Fig. 2.1
Molecular pathology of field cancerization in HNC
Because HNSCC has well-established premalignant and precursor lesions, and molecular pathologic progression model, the ability to use biomarkers to detect this cancer early should be easy. A noninvasive test to detect the early precursor lesions targeting molecular genetic alterations should enable early detection and deployment of possible curative interventions. Works at developing such tests using saliva or blood are being explored.
2.4 Molecular Pathology of HNC
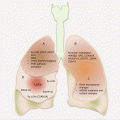
Head and neck cancers can originate in any tissue of the head and neck region. However, excluding salivary gland tumors, almost all HNSCCs are from three anatomic regions, the oral cavity, larynx, and the pharynx (Fig. 2.2). Not only is this important for disease management, but this also raises the possibility of early noninvasive detection of many HNCs using oral fluids or blood.
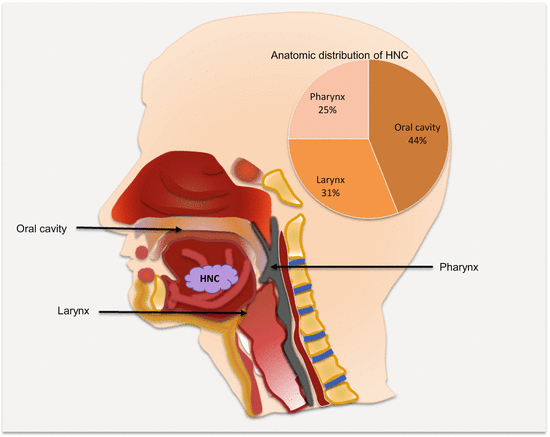

Fig. 2.2
Frequencies and common anatomic locations of HNC
2.4.1 Molecular Classification of HNC
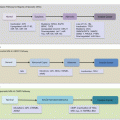
Head and neck cancer has two major etiologic factors, toxins (alcohol and tobacco toxicity) and viral infections. Epstein–Barr viral infection is a risk factor for nasopharyngeal (NPC), while HPV infection causes a subset of HNSCCs. The HPV-negative HNSCCs are associated with tobacco and alcohol use. These subclasses have distinct molecular genetic changes, clinical course, and prognosis. For example, HPV-positive tumors harbor wild-type TP53 and have favorable outcomes. It is conceivable that the HPV-negative tumors will be genetically homogenous. However, work by Smeets and colleagues suggests otherwise [1]. Using array CGH, they uncovered three genetically distinct groups of HPV-negative tumors with prognostic relevance. Tumors in “group 1” had hardly any chromosomal instability (CIN) and were associated with wild-type TP53, female gender, and nonalcoholics. An intermediate group (group 2) harbored high chromosomal aberrations, while at the extreme end were those with very high levels of CIN (group 3). Patients in “group 1” had the best prognosis, with the worse being among “group 3” patients. On the basis of this and other data, Leemans and colleagues dichotomized HPV-negative tumors into those with high and low CIN (Fig. 2.3) [2]. Tumors with low CIN, which are in the minority (15 %), harbor wild-type TP53, are near diploid, and hence are associated with good outcomes. Those with high CIN are mostly aneuploid with TP53 mutations and are associated with poor prognosis.
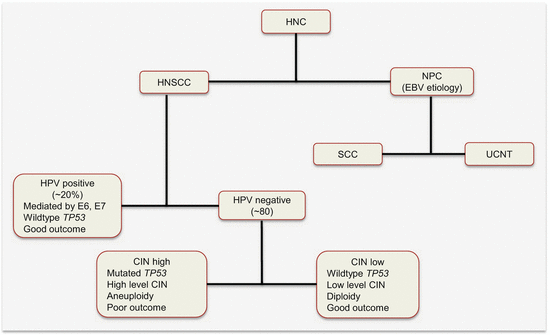

Fig. 2.3
Etiologic classification of HNC with some associated molecular alterations and prognosis. HNSCC head and neck squamous cell carcinoma, NPC nasopharyngeal carcinoma, EBV Epstein–Barr virus, SCC squamous cell carcinoma, UCNT undifferentiated carcinomas of the nasopharyngeal type
An earlier study by Chung et al. using gene expression signatures uncovered four distinct molecular subtypes of HNSCC, also exhibiting different prognosis [3]. These molecular subtypes included tumors enriched for EGFR pathway, mesenchymal cell, normal epithelial-like cell, and high antioxidant enzyme gene signatures. The worse prognosis was among tumors with EGFR pathway gene expression signature.
2.4.2 Molecular Pathology of HPV-Positive HNC
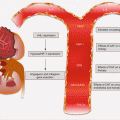
High-risk HPV (types 16 and 18) infections of the oropharynx account for ~20 % of all HNSCCs. These cancers are distinct from non-HPV HNSCC, especially at the molecular level. They are associated with wild-type TP53 genotype, have a favorable outcome, and are mostly confined to the oropharynx. Indeed over 50 % of all oropharyngeal squamous cell carcinomas are HPV positive. In addition to the upper aerodigestive tract, oncogenic HPV also accounts for the majority of cervical cancers, and this epithelial cellular transformation is mostly mediated by viral oncoproteins E5, E6, and E7. These proteins primarily target and deregulate members of the cell cycle and other oncogenic signaling pathways (Fig. 2.4). For example, E5 oncoprotein can interfere with the internalization and destruction of EGFR leading to increased pathway activity. Also the ubiquitin ligase, E6 oncoprotein, targets and degrades p53 tumor suppressor protein to prevent apoptosis, while E7 binds to and inhibits the activity of RB in sequestering E2F to promote cycle progression. Disruption of p53 and RB functions will cause cell cycle progression without mitogen activation. E7 can also promote cell cycle activity by inhibiting CDK inhibitors including p21 and p27.
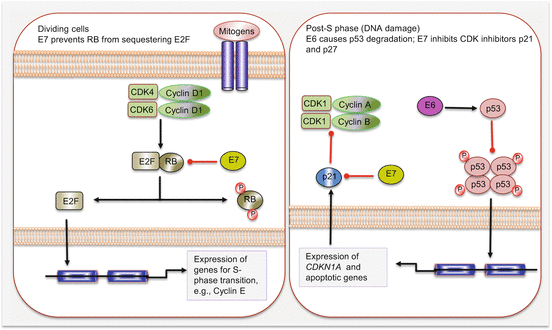

Fig. 2.4
Cell cycle deregulation by HPV E6 and E7 oncoproteins. Red lines with blunt ends indicate inhibitory pathways
2.4.3 Specific Genetic Alterations in HNC
Head and neck cancer is genetically characterized by abnormalities at almost all chromosomal regions, many of which harbor critical oncogenes and tumor suppressor genes. Indeed, the only chromosomes not involved in head and neck carcinogenesis are 12 and 16. The chromosomal changes include amplifications and gains at oncogenic loci, 3q26 (PIK3CA), 7p11.2 (EGFR), 11q13 (CCND1), and 7q31 (MET), as well as losses or homozygous deletions of tumor suppressor gene loci including 9p21 (CDKN2A), 18q21 (SMAD4), 10q23 (PTEN), and 17p13 (TP53). Together with mutations and epigenetic alterations, the functions of several genes are altered, which eventually reflect on the activities of several carcinogenic signaling pathways including the PI3K/AKT, RAS-MAPK, EGFR, TGFR, p53/RB, and possibly NF-κB pathways.
2.4.3.1 EGFR Alterations in HNC
Receptor tyrosine kinase (RTK) signaling responds to growth factor signals to trigger and intergrade the RAS-MAPK and PI3K/AKT pathways that control tumor growth and survival. One of the well-studied RTKs is the ERBB family. The four family members can undergo ligand-mediated homodimerization or heterodimerization to activate the EGFR pathway. Downstream signaling cascade includes the PI3K, RAS-MAPK, phospholipase C (PLC), and JAK/STAT pathways. Additionally, activated EGFR can translocate into the nucleus and either act as a transcription factor for genes such as CCND1 or transcriptional coactivator for STAT and some miRNAs. EGFR is oncogenic in HNSCC, being amplified in up to 30 % of HNCs. The gene is also mutated and overexpressed in some HNCs. The elevated levels and mutations of EGFR in theses tumors can cause constitutive signaling via spontaneous monomeric receptor dimerization and tyrosine kinase activation. Co-expression of both receptor and ligand (TGFα) indicates it can cause autocrine signaling in some cancer cells. EGFR variant 3 mutant (EGFRvIII—lacks exons 2–7) is expressed in ~42 % of HNCs and is also associated with lack of response to anti-EGFR monoclonal antibody, cetuximab biotherapy. EGFR overexpression is a prognostic factor, conferring poor outcomes in HNC patients.
2.4.3.2 TGFβ Pathway Alterations in HNC
Transforming growth factor β (TGFβ) is a growth inhibitory signaling pathway associated with HNSCC. The TGFβ1 ligand interacts with TGFβ receptors to phosphorylate and activate SMAD2 and SMAD3, which then combine with SMAD4 to form the SMAD complex. The SMAD complex enters the nucleus to bind transcription factors, coactivators, or corepressors to control expression of TGFβ1 pathway target genes. Pathway activation is associated with decreased cell proliferation, decreased cell survival, and increased apoptosis. The SMAD and TGFβRII locus on chromosome 18q are frequently lost in HNC fields and contribute to invasive cancer formation. Additionally, TGFβ receptors are downregulated in HNCs.
2.4.3.3 PI3K/AKT Pathway Alterations in HNC
PIK3CA encodes p110α, a catalytic subunit for the PI3K class Ia molecules. The locus of PIK3CA, chromosome 3p26, is commonly gained or amplified in HNCs. Additionally, activating mutations in PIK3CA occur in 10–20 % of HNCs, especially HPV-negative tumors, and these cancers tend to have increased vascular invasion and lymph node metastasis. The PI3K pathway negative regulator, PTEN, is inactivated in ~10 % of HNCs as well.
2.4.3.4 TP53 Alterations in HNC
Tumor protein p53 (TP53) and retinoblastoma (RB) are commonly mutated in HNSCC, and the proteins are targets of viral oncoproteins E6 and E7. These alterations lead to cell cycle deregulation, enabling cells to escape senescence and hence replicate uncontrollably. Besides its role in apoptosis, p53 controls the cell cycle at G2, following DNA synthesis at the S phase. P53 primarily functions to prevent expansion of unrepaired replicative errors, should they occur. In normal proliferating cells, p53 is usually targeted by MDM2 for ubiquitin-mediated proteasomal degradation, thus keeping the levels very low. In conditions of DNA damage, the p53 pathway is activated leading to increased expression and activity of p21CIP (CDKN1A), which stops cell cycle progression by negatively controlling the activity of cyclin/CDK complexes. Somatic mutations in TP53 occur in 60–80 % of HNCs, mostly HPV-negative tumors. In HPV16-infected HNCs, the E6 oncoprotein targets and inactivates p53.
2.4.3.5 Cell Cycle Gene Alterations in HNC
Cell cycle progression through the G1 to S phase is controlled by RB tumor suppressor protein. In non-proliferating cells, RB binds and inactivates E2F transcription factor. In proliferating cells, activated cyclin D1/CDK4 and cyclin D1/CDK6 complexes phosphorylate RB leading to the release of E2F, which targets and induces the expression of genes including cyclin E for G1–S phase transition. Cyclin E/CDK2 complex further phosphorylates RB, driving cell cycle progression into the S phase. In order for senescence and cell differentiation to occur, p16 is expressed, and this inhibits the cyclin D1/CDK4 and cyclin D1/CDK6 complexes. CCND1 is amplified or gained in over 80 % of HNSCCs and homozygous loss at chromosome 9p21, the CDKN2A locus is frequent in HNCs. Gene mutations and promoter hypermethylation also inactivate CDKN2A. HPV oncoprotein E7 targets the RB pocket proteins RB1, RBL1 (p107), and RBL2 (p130).
2.4.3.6 MET Alterations in HNC
Mesenchymal-epithelial transition factor (MET), located on chromosome 7p31, is a receptor for hepatocyte growth factor or scatter factor. It encodes an RTK that upon ligand interaction activates the PI3K and RAS-MAPK pathways. Mutations and amplifications of MET are found in HNSCC. MET activation is associated with increased cell growth, angiogenesis, and the metastatic phenotype.
2.4.3.7 Telomere Alterations in HNC
Another important alteration in HNC is telomere lengths. Telomerase (TERT) expression is increased in ~80 % of HNCs, but the TERT locus on chromosome 5p15.33 is not frequently gained or amplified in HNCs.
2.5 Circulating HNC Biomarkers
The clinical potential of ccfDNA and epigenetic alterations in ctDNA, miRNA, and serum proteins has been explored as HNC biomarkers. Circulating HNC cells demonstrate some utility in disease staging, prognosis, and treatment predictions. Validation of these liquid biopsy biomarkers will augment HNC management.
2.5.1 Circulating Cell-Free Nucleic Acids as HNC Biomarkers
Nucleic acid integrity is compromised in circulation of HNC patients, enabling their exploration for disease detection. For example, a study that employed multivariate analysis, plasma DNA integrity index (DII) was found to be significantly higher in patients with HNC than in controls. Optimal performance was obtained at a sensitivity of 84.5 % and a specificity of 83 % using a DII cutoff value of 0.82. But, this study failed to observe changes in postoperative samples as will be expected if the differences were due to cancer origin of high molecular weight DNA. The authors asserted to the possibility of residual population of cells with altered DNA degradation despite surgical removal of the cancer. Given the established importance of field cancerization in HNC, this conclusion is conceivable for this type of cancer. DII may only be useful for diagnostic purposes [4]. Chan et al. also examined DII before and after curative radiotherapy and compared them to controls without cancer [5]. They targeted LEP (leptin gene) fragments of lengths 105 bp and 201 bp. DII, defined as the ratio of 201 bp to 105 bp DNA fragments, was significantly higher in plasma from nasopharyngeal carcinoma (NPC) patients than controls. After radiotherapy, DII was reduced in 70 % of the cancer patients. Patients with lack of reductions in DII had significantly poorer survival revealed by Kaplan–Meier analysis. Persistently high DII was associated with significant poor disease-free survival (DFS). Another investigation targeted RNA alterations in HNC, dubbed RNA integrity index (RII). Because of increased RNAase activity in circulation of cancer patients, reduced RNA integrity in plasma of cancer patients is expected. In this study, RII, defined as the ratio of 3′-to-5′ transcripts of GAPDH, was assayed as a diagnostic and predictive biomarker of patients with NPC. RII was significantly lower in plasma from patients with untreated NPC compared to healthy controls, and this ratio correlated with tumor stage. Interestingly, 74 % of the patients exhibited significant increased plasma RII after radiotherapy [6]. Thus, DII or RII shows potential as biomarkers of HNC, but the lack of specificity to this cancer, except when used in high-risk individuals, may hamper generalized utility.
2.5.2 Circulating HNC Epigenetic Biomarkers
Head and neck cancer is characterized by early epigenetic alterations (e.g., CDKN2A promoter hypermethylation). Several of these epigenetic changes have been explored in circulation, though disproportionately focused on NPC patients.
Methylation of CDKN2A, MGMT, GSTP1, and DAPK was assayed in primary HNCs and then in paired sera. Except GSTP1, 55 % of tumors had promoter hypermethylation in at least one of the genes. Fifty patients had paired sera, and methylation was demonstrated in 42 % of these samples. DAPK methylation was associated with lymph node metastasis and advanced disease state [7]. Methylated DAPK as a diagnostic biomarker in NPC has been assayed in tumor tissues, cell lines, plasma, and buffy coat samples. Hypermethylation was observed in 75 % of NPC tissues and 80 % of cell lines. Fifty percent (50 %) of plasma samples and 25 % of buffy coat samples harbored DAPK methylation, with 66.7 % of either sample being positive. DAPK methylation was not stage dependent and therefore can serve as an early detection biomarker [8]. Hypermethylation of high in normal 1 (HIN-1) was also assayed in primary NPC tissues and matched nasopharyngeal swabs, throat rinses, and blood samples. Methylation in association with gene downregulation was present in all cell lines and 77 % of primary NPC samples. Methylation was in 46 %, 19 %, 18 %, and 46 % of nasopharyngeal swabs, throat rinses, plasma, and buffy coat, respectively [9]. This same group examined the possible use of gene methylation as screening and predictive biomarkers. Methylation of CDH1, DAPK, CDKN2B, CDKN2A, and RASSF1A was present in 71 % of plasma from NPC patients. Methylation of at least one of CDH1, DAPK, or CDKN2A was in 38 % of recurrences but none of the patients in remission [10]. The screening potential of RIZ1 methylation has been demonstrated. RIZ1 methylation in primary NPC tissues, cell lines, body fluids, and swabs was examined. Hypermethylation was present in cell lines, 60 % of primary NPCs, 37 % of nasopharyngeal swabs, 30 % of mouth or throat washes, 23 % of plasma, and 10 % of buffy coat samples. RIZ1 has a role for screening for NPC because all controls were negative (no false-positive results—100 % specificity as in many methylation assays) [11].
Global methylation studies have identified risk and possible early detection methylation patterns and biomarkers in circulation of patients with HNC [12, 13]. In a case–control study, Hsiung et al. reported that DNA hypomethylation in blood was significantly associated with elevated risk of HNSCC. A 1.6-fold risk was demonstrated even when controlled for other HNSCC risk factors. Smokers, individuals with low folate intake, and those with MTHFR genotypes had decreased global methylation and hence increased risk of cancer. In another genome-wide methylation profiling, six CpG islands methylated in circulation were most useful in HNC early detection. Methylation of FGD4, SERPINF1, WDR39, IL27, HYAL2, and PLEKHA6 achieved an AUROCC of 0.73 (95 % CI 0.76–0.92) in HNC detection.
2.5.3 Circulating HNC Noncoding RNA Biomarkers
Oncomirs and tumor suppressormirs from HNC have been studied and multiple relevant targets identified. These miRNA and target deregulation, probably the tip of the iceberg at the moment, influence important signaling pathways involved in HNC progression.
2.5.3.1 Oncomirs and Tumor Suppressormirs in HNC
Overexpressed in HNC and with identified targets are miR-21, miR-17-92 polycistron, miR-181b, miR-106a, miR-106b-92, miR-106b-25 cluster, miR-155, miR-205, miR-221, and miR-345. The elevated level of miR-21 is associated with increased cell growth and apoptotic suppression via reduced cytochrome c release. These functions are achieved by suppression of a number of tumor suppressors including PTEN, PDCD4, TPM1, and SERPINB5. Indeed, overexpression of miRNAs including miR-21, miR-181b, and miR-345 drives leukoplakia toward invasive OSCC. In addition to miR-21, PTEN is a target of miR-205 in HNSCC.
Cell cycle deregulation is an important event in HNSCC progression. In addition to epigenetic and genetic alterations, cell cycle components are major alterations in head and neck carcinogenesis. For instance, miR-221 suppresses CDKN1B and CDKN1C mRNAs, while miR-17-92 and miR-106a target and degrade RB1. MiR-106a, 106b-25, and miR-17-92 clusters target CIP cyclin/CDK inhibitor, CDKN1A, that encodes p21. Consistently, knockdown of miR-106b-25 in HNSCC cells decreases cell proliferation via G1 phase arrest. Additionally, miR-106b-92 and miR-17-92 cluster interfere with TGFβ and MYC signaling pathway communication leading to cell cycle deregulation and loss of apoptotic response. MiR-155 targets APC tumor suppressor gene and also controls TGFβ-mediated epithelial-to-mesenchymal transition by targeting RHOA transcripts.
Tumor suppressor miRNAs identified so far in HNSCC include let-7, miR-100, miR-125a/b, miR-133, and miR-200a. All let-7 family members (except let-7i) are downregulated in HNSCC, and these target KRAS and HMGA2. MiR-125a/b degrades ERBB2. Thus, the loss of miR-125a/b increases ERBB2 expression associated with EGFR signaling in HNSCC. Downregulation of miR-100 in HNSCC leads to overexpression of oncogenes such as ID1, FGFR1, and MMP13. Pyruvate kinase M2 (PKM2), a well-known metabolic regulator in cancer cells, is a target of miR-133a/b. MiR-200a, which is downregulated in OSCC, targets E-cadherin repressors ZEB1 and ZEB2, thus promoting epithelial-to-mesenchymal transition, tumor cell migration, and invasion.
2.5.3.2 Circulating HNC miRNA Biomarkers
A number of miRNAs have been examined in circulation of patients with HNSCC as diagnostic and prognostic biomarkers. Five circulating miRNAs, miR-16, let-7b, miR-338-3p, miR-223, and miR-29a, yielded an AUROCC of >0.80, suggesting their potential utility as noninvasive biomarkers for the detection of oral cancer or high-grade lesions [14]. MiR-17, miR-20a, miR-29c, and miR-223 were of diagnostic relevance for NPC [15]. Plasma miR-31 levels are much higher in OSCC patients than controls, and the levels decreased after surgical tumor removal. Also plasma miR-27b levels are reduced in oral cancer patients. In patients with oral cancer and precancerous lesions, plasma mR-196a and miR-196b levels were significantly higher than controls. While both demonstrated excellent performances independently, the combination of the two yielded an AUROCC of 0.845 for detection of precancerous lesions and 0.963 for oral cancer [16].
Circulating prognostic HNC biomarkers include miR-21, miR-26b, and miR-181. The expression of miR-181 is associated with progression of leukoplakia to invasive OSCC. Circulating and tissue miR-181 levels correlate with lymph node metastasis, invasiveness, and overall poor survival. This miRNA promotes cell migration and invasion [17]. MiR-21 (established to be markedly upregulated in cancer tissues) is significantly increased in plasma of patients as well. Plasma concentrations of miR-21 and miR-26b reduced postoperatively in patients with good prognosis but remained high even after surgery in those with poor outcome [18].
2.5.4 Circulating HNC Serum Protein Biomarkers
There are several altered proteins in circulation of HNC patients. Mostly ELISA has been used to evaluate this extensive number of serum biomarkers as diagnostic and prognostic biomarkers of HNSCC. However, only a few appear as being clinically useful. The comprehensive meta-analytical synthesis by Guerra et al. [19] reveals the following:
Majority of the studies involved single biomarkers, which expectedly are associated with dismal performances.
Panel biomarkers had improved sensitivity and specificity for HNSCC detection.
Thus, 34.3 % of panel and 12.8 % of single serum biomarkers examined were discriminatory for HNSCC.
Commonly investigated serum biomarkers were CYFRA21-1, SCCA, and CEA.
Of 15 single biomarkers, prolactin, catalase, glutathione, and β2-microglobin were of superior diagnostic performances for HNC. But β2-microglobin appears to be the most valid single biomarker with diagnostic potential.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree