© Springer International Publishing 2015
Primo N. Lara and Eric Jonasch (eds.)Kidney Cancer10.1007/978-3-319-17903-2_55. Genetic Heterogeneity of Kidney Cancer
(1)
Deapartment of Internal Medicine, Division of Hematology-Oncology, University of California Davis Comprehensive Cancer Center, Sacramento, CA, USA
(2)
Deapartment of Internal Medicine, Division of Hematology-Oncology, University of California Davis School of Medicine, UC Davis Comprehensive Cancer Center, Sacramento, CA, USA
(3)
Department of Genitourinary Medical Oncology, UT MD Anderson Cancer Center, Houston, TX, USA
Key Points
The observation that renal cell cancer patients often develop mixed responses to therapy has led to the hypothesis that intratumoral heterogeneity may exist within individual patient tumors.
Advances in molecular phenotyping techniques have led to the identification of significant intratumoral genetic heterogeneity in renal cell cancers of clear cell and variant histologies.
Phylogenetic trees constructed by inferring ancestral relationships of tumor subclones demonstrate branched rather than linear evolution patterns in individual renal cell cancers. The majority of known driver mutations in renal cell carcinoma map to branching and not to truncal portions of phylogenetic tree constructions.
Individual renal cell cancers demonstrate evidence of convergent phenotypic evolution by tumor subclones. SETD2 and other tumor suppressor genes have undergone distinct genetic alterations in multiple spatially separated regions within a single tumor converging on loss of function.
Intratumoral heterogeneity in renal cell cancer may confound clinical decision-making on therapeutic strategies, alter drug development strategies, and may require the identification of improved biomarkers to guide clinical practice.
5.1 Background
Clinicians have long suspected that significant heterogeneity may exist within individual tumors and their metastases [1]. Patients with metastatic renal cell cancer (RCC) are known to develop mixed responses to therapy suggesting the presence of tumor subclones and clonal selection [1, 2]. In the past, traditional laboratory techniques were employed to gain insights into the molecular basis of such heterogeneity. For example, chromosomal analysis has shown that a more complex cytogenetic pattern is found in more aggressive and advanced RCC, suggesting that sequential accumulation of chromosome changes may play a role in cancer progression [3]. An evaluation of chromosomal mutations and mitotic segregation patterns in RCC showed that in a subset of tumors, there were abnormally shortened telomere repeat sequences, chromosomal breakage-fusion-bridge events, multipolar configurations, and supernumerary centrosomes [4]. These observations suggested that changes in cell division machinery may be involved in the evolution of complex karyotypes and genetic intratumoral heterogeneity in a subgroup of RCC. Furthermore, Ljundberg et al. employed flow cytometry to evaluate DNA ploidy in 200 consecutive RCC specimens: these investigators reported that there was frequent heterogeneity in these specimens and concluded that “multiple samples must be investigated to evaluate properly the malignant character of renal cell carcinoma” [5]. Early investigations into the metastatic heterogeneity of RCC also involved the development of a nude mouse model for evaluating RCC metastasis [6]. Employing this mouse model, Fidler and colleagues used the SN12C RCC line which had a heterogeneous subpopulations of cells with varied metastatic potential, as well as cells derived from spontaneous lung metastases [6]. These investigations provided some early tools to individually study RCC variants with high metastatic potential and to develop models for dissecting tumor evolution and metastasis [7].
More recently, advances in molecular phenotyping techniques such as next-generation sequencing have allowed for a deeper understanding of RCC evolutionary biology through the detection of genetically distinct subclones within individual tumors and the characterization of clonal architecture [8]. This technology has subsequently been used to study intratumor heterogeneity not just in RCC but in a diverse range of tumor types including breast cancer [9, 10], pancreatic cancer [11], ovarian carcinoma [11], and acute leukemia, [13–15], among others. This chapter will summarize recent data that employed modern molecular techniques to shed light on RCC heterogeneity, clonal evolution, and the potential clinical implications of these findings.
5.2 Intratumor Heterogeneity
Employing whole-exome sequencing to study intratumoral heterogeneity, Gerlinger et al. analyzed multiple regions from ten primary tumors and their associated metastases in three cases [16, 17]. These investigators found that 67 % of identified somatic mutations were heterogeneous and not detectable across all sampled regions within an individual tumor. Mutational intratumoral heterogeneity was seen for multiple tumor suppressor genes converging on loss of function. In addition, these investigators applied a 110-gene signature shown to classify ccRCC into good prognostic and poor prognostic molecular subgroups on spatially distinct regions of one tumor sample. The metastatic tumors and one region of the primary tumor segregated into the good prognostic subgroup, while the remaining regions of the primary tumor segregated into the poor prognostic subgroup, further illustrating the significant molecular heterogeneity within an individual tumor.
Martinez et al. [18] further characterized the extent of intratumoral heterogeneity by comparing individual tumor samples of clear cell RCC with unrelated tumor samples collected from the Cancer Genome Atlas (TCGA). Twenty-five percent of tumor biopsies demonstrated greater genetic similarity with unrelated tumor samples than with samples originating from the same primary tumor.
To further assess intratumoral genetics that underlie the mutational spectrum of clear cell RCC, Xu et al. performed single-cell exome sequencing using material from a kidney cancer and its adjacent normal kidney tissue [19]. These investigations revealed that the kidney tumor was unlikely to have evolved from mutations in VHL and PBRM1. Quantitative population genetic analysis interestingly showed that the tumor did not contain any significant clonal subpopulations. However, this analysis revealed that mutations with different allele frequencies within the population had different mutational spectra, suggesting that clear cell RCC “may be more genetically complex than previously thought” [19]. Novel algorithms to construct phylogenetic models of tumor progression at the cellular level – incorporating copy number changes at the scale of single genes, entire chromosomes, and the whole genome – are currently under development and may help shed additional light on the implications of single-cell sequencing [20].
5.3 Heterogeneity in Variant RCC Histologies
Investigations of RCC heterogeneity extend beyond that of clear cell histology into that of less common variant subtypes. Using next-generation sequencing (NGS), Durinck et al. analyzed exome, transcriptome, and copy number alteration data from 167 primary human tumors that included renal oncocytomas and non-clear cell RCC consisting of papillary (pRCC), chromophobe (chRCC), and translocation (tRCC) subtypes [21]. Within the non-clear cell subtypes, these investigators found that pRCCs had a higher mutation rate than chRCCs and renal oncocytomas and that genes altered in non-clear cell RCC were distinct from that reported with clear cell histology. Ten significantly mutated genes were identified in pRCC, including MET, NF2, SLC5A3, PNKD, and CPQ. In chRCC, the following genes were found to be significantly mutated: TP53, PTEN, FAAH2, PDHB, PDXDC1, and ZNF765. Interestingly, gene expression analysis identified a five-gene set that molecularly classified chRCC, renal oncocytoma, and pRCC.
Malouf et al. described the genomic and epigenetic characteristics of translocation renal cell carcinoma (tRCC), a rare subtype of kidney cancer involving the TFEB/TFE3 genes [22]. These investigators reported moderate cytogenetic heterogeneity in this rare tumor type, with 31.2 % and 18.7 % of cases presenting similarities with clear cell and pRCC profiles, respectively. The most common alterations seen were 17q gain in 44 % and 9p loss in 37 %. Exome sequencing of tRCC revealed a distinct mutational spectrum with frequent mutations in chromatin-remodeling genes [23].
A study of molecular heterogeneity in RCC with sarcomatoid differentiation using X-chromosome inactivation analysis suggested that both clear cell and sarcomatoid components of renal cell carcinomas were derived from the same progenitor cell [24]. Additionally, different patterns of allelic loss in multiple chromosomal regions were reported in clear cell and sarcomatoid elements from the same patient, suggesting divergence during RCC clonal evolution.
5.4 Branching Evolution
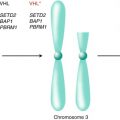
Gerlinger et al. utilized genetic analyses to construct phylogenetic trees by inferring ancestral relationships of tumor subclones [17]. These phylogenetic trees of ccRCC demonstrated branched rather than linear evolutionary patterns in all ten samples analyzed. Early ubiquitous genetic alterations were mapped to the truncal portion of the phylogenetic trees, while later heterogeneous alterations occurring in separate spatial regions composed the branches. Known driver mutations of ccRCC were mapped onto the phylogenetic trees to determine whether specific driver genes were predominantly altered on truncal or branch portions [25]. Alterations in the von Hippel-Lindau (VHL) tumor suppressor gene were identified ubiquitously on the truncal portions of each phylogenetic tree consistent with its role as a critical founder event in the pathogenesis of ccRCC. However, the majority of known driver mutations were mapped onto the branches of the phylogenetic trees with 73 % of driver mutations identified in subclonal populations. These mutations included alterations in PTEN, SETD2, KDM5C, PBRM1, and BAP1 expression identified in spatially separate subclones.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree