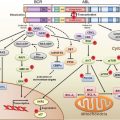
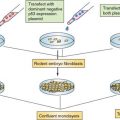
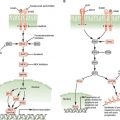
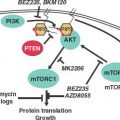
EXAMPLES OF GENETIC CHANGES TO FACILITATE TREATMENT DECISIONS

SUMMARY
The understanding of the genetic and genomic differences among individuals that affect cancer susceptibility and the genetic and genomic changes that are critical for the initiation and progression of cancer is increasing at a rapid pace. The rapid decrease in the cost of DNA sequencing and other genomic analysis is fueling this increase in knowledge. This new knowledge is helping in accurate prediction, early detection, and prognosis of cancer. Genetic differences among individuals and somatic changes during the development of cancer are also important in determining the appropriate treatment strategies for each patient. The use of genetic and genomic information is referred to as personalized medicine, which has the ability to transform the practice of oncology.
Selected References
The full list of references for this chapter appears in the online version.
1. Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 2000;343:78.
2. Ashley DJ. Oesophageal cancer in Wales. J Med Genet 1969;6:70.
3. St. John DJ, McDermott FT, Hopper JL, et al. Cancer risk in relatives of patients with common colorectal cancer. Ann Intern Med 1993;118:785.
4. Nishisho I, Nakamura Y, Miyoshi Y, et al. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science 1991;253:665.
5. Kinzler KW, Nilbert MC, Su LK, et al. Identification of FAP locus genes from chromosome 5q21. Science 1991;253(5020):661.
6. Nakamura Y, Nishisho I, Kinzler KW, et al. Mutations of the adenomatous polyposis coli gene in familial polyposis coli patients and sporadic colorectal tumors. Princess Takamatsu Symp 1991;22:285.
7. Lynch HT, Krush AJ. Heredity and adenocarcinoma of the colon. Gastroenterology 1967;53:517.
8. Parsons R, Li GM, Longley MJ, et al. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell 1993; 75:1227.
9. Aaltonen LA, Peltomäki P, Leach FS, et al. Clues to the pathogenesis of familial colorectal cancer. Science 1993;260:812.
10. Powell SM, Zilz N, Beazer-Barclay Y, et al. APC mutations occur early during colorectal tumorigenesis. Nature 1992; 359:235.
11. Leach FS, Nicolaides NC, Papadopoulos N, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 1993;75:1215.
12. Fishel R, Lescoe MK, Rao MR, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 1993;75:1027.
13. Papadopoulos N, Nicolaides NC, Wei YF, et al. Mutation of a MutL homolog in hereditary colon cancer. Science 1994;263:1625.
14. Bronner CE, Baker SM, Morrison PT, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature 1994;368:258.
15. Nicolaides NC, Papadopoulos N, Liu B, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 1994;371:75.
16. Miyaki M, Konishi M, Tanaka K, et al. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet 1997;17:271.
17. Lipkin SM, Wang V, Jacoby R, et al. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat Genet 2000;24:27.
18. Loukola A, Vilkki S, Singh J, Launonen V, Aaltonen LA. Germline and somatic mutation analysis of MLH3 in MSI-positive colorectal cancer. Am J Pathol 2000;157:347.
19. Aaltonen LA, Salovaara R, Kristo P, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med 1998;338:1481.
20. Loukola A, Salovaara R, Kristo P, et al. Microsatellite instability in adenomas as a marker for hereditary nonpolyposis colorectal cancer. Am J Pathol 1998;155:1849.
21. Salovaara R, Loukola A, Kristo P, et al. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol 2000;18:2193.
22. de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer 2004;4:769.
23. Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet 2007;39:989.
24. Al Olama AA, Kote-Jarai Z, Giles GG, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet 2009;41:1058.
25. Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet 2009;41:1122.
26. Tenesa A, Farrington SM, Prendergast JG, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet 2008;40:631.
27. Pomerantz MM, Ahmadiyeh N, Jia L, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 2009;41:882.
28. Ahmadiyeh N, Pomerantz MM, Grisanzio C, et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. 2010 Proc Natl Acad Sci U S A 2010;107:9742.
29. Hall JM, Lee MK, Newman B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 1990; 250:1684.
30. Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994;266:66.
31. Futreal PA, Liu Q, Shattuck-Eidens D, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science 1994;266:120.
32. Faca VM, Song KS, Wang H, et al. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med 2008;5:e123.
33. Pitteri SJ, JeBailey L, Faça VM, et al. Integrated proteomic analysis of human cancer cells and plasma from tumor bearing mice for ovarian cancer biomarker discovery. PLoS One 2009;4:e7916.
34. Hung KE, Faça V, Song K, et al. Comprehensive proteome analysis of an Apc mouse model uncovers proteins associated with intestinal tumorigenesis. Cancer Prev Res 2009;2:224.
35. Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007;450(7173):1235.
37. Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 2007;7(4):233.
38. Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561.
39. Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 2007;448:595.
40. Dave SS, Fu K, Wright GW, et al. Lymphoma/Leukemia Molecular Profiling Project. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med 2006;354:2431.
41. Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351(27):2817.
42. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061.
43. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008; 321:1807.
44. Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321:1801.
45. Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 2010;463:191.
46. Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 2009;361:1058.
47. Nowell P, Hungerford D. A minute chromosome in human chronic granulocytic leukemia Science 1960;132:1497.
48. Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukemia. Nature 1973;243:290.
49. Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med 2001;344:1031.
50. Druker BJ, Sawyers CL, Kantarjian H, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med 2001;344:1038.
51. Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344:783.
52. Arteaga CL. ErbB-targeted therapeutic approaches in human cancer. Exp Cell Res 2003;284:122.
53. Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497.
54. Lynch TJ, Bell DW, Sordella R, et al. activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129.
56. Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947.
61. Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757.
63. Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 2010;363:809.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree