Fig. 8.1
(a) Fusion probe FISH assay negative for translocation (“split signal”). (b) Fusion probe FISH assay positive for translocation (“fusion signal”)
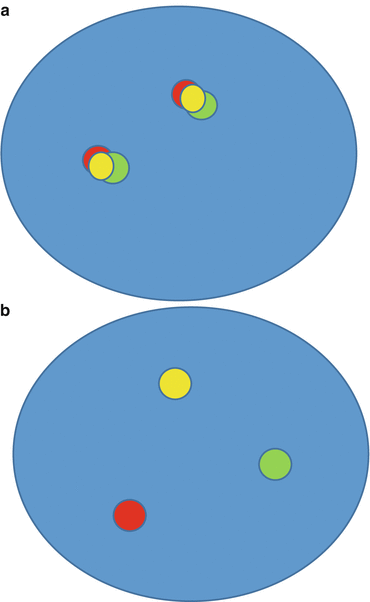
Fig. 8.2
(a) Break-apart probe FISH assay negative for translocation (“fusion signal”). (b) Break-apart probe FISH assay positive for translocation (“split signal”)
Interpretation
The identification of non-overlapping cells/nuclei is the first step in the interpretation . It is essential that a pathologist identify adequate tumor areas on the routine H&E slide that is adjacent to section submitted for FISH analysis. An experienced cytotechnologist who has undergone specific FISH training in solid tumors should analyze the slides. Interpretation should be performed in areas of the slide with good signal, in which at least 50% of all nuclei are easily analyzable, with minimal background and autofluorescence. The FISH signal intensity should be consistently greater than background intensity.
Common Clinical Assays for Gene Rearrangements in Lung Cancer
ALK Gene Rearrangement
FISH Probe Design
The most common of ALK rearrangements involve a pericentric inversion on the short arm of chromosome 2, inv. [2] (p21p23), which creates a fusion gene encoding the amino-terminal portion of EML4 (2p21) and the intracellular region of ALK (2p23), genes that are normally approximately 13 Mb apart [2]. Although the EML4-ALK fusion is the most common, other less common variant fusions have been reported, including translocations with other chromosomes (KIF5B-ALK, TFG-ALK) [3, 4]. The Vysis LSI ALK break-apart FISH probe kit (Abbott Molecular) was used to identify patients with ALK rearrangement positive NSCLC in the first clinical trials with ALK inhibitors, and therefore, the US Food and Drug Administration (FDA) approved this commercially available assay as a companion diagnostics for detection of ALK rearrangements in lung cancer [5]. The break-apart probe is designed by labeling the 3′(telomeric) 300-kb part fusion breakpoint with orange fluorochrome (Spectrum Orange, often referred as red) and the 5′ 442′-kb probe by a green signal (SpectrumGreen) (Fig. 8.3).
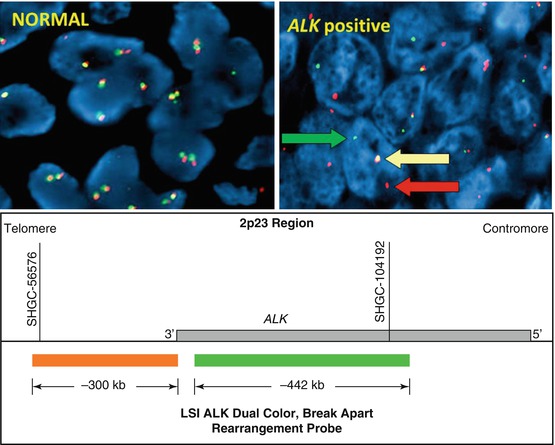

Fig. 8.3
Break-apart ALK FISH probe
Interpretation
The interpretation of ALK FISH assay may be challenging because the 5′ and 3′ probes are genetically very close [6–9]. Therefore, cells with normal pattern show fused signals. For the same reason in some cases split signal can be very narrow that the signals seems to be very close and fused in otherwise ALK rearranged cells. In the interpretation of ALK FISH, it is essential to pay attention to distance between the signals which should measure at least two signal diameters in cases positive for translocation. Any distance that is less than two signal diameter is considered to represent lack of gene rearrangement. A minimum of 50 tumor cells should be scored if there is one scorer, and a minimum of 100 tumor cells is needed if there are two scorers.
The assay is considered to be positive for ALK rearrangement if at least 15% of tumor cells show rearrangement. FISH patterns that are considered to represent gene rearrangement include split pattern and isolated 3′ pattern (Fig. 8.4). The number of accompanied fused 5′-3′ signals in the cell is not important for pattern classification. Isolated 5′ pattern may also be identified and is considered to represent nonfunctional reciprocal fusion product. Although this pattern has been reported to be associated with a rare BIRC6-ALK fusion, it should not be interpreted as rearrangement positive [10, 11]. FISH criteria particularly in respect to isolated 3′ (“single orange”) have been recently challenged [12, 13]. Cases classified as rearrangement positive based on isolated 3′ showed a higher rate of fusion negative cases by NGS and IHC than the group with a FISH split signal indicating that these cases may be FISH false positive [13, 14]. Isolated 3′ may be a result of technical factors such as nuclear sectioning causing loss of the 5′ (green) probe binding site, or simply observer error. Technical errors can’t be reliably excluded in a case with a lower percentage of nuclei positive for rearrangement. Overall, cases with atypical signal patterns should be tested by another method such as IHC or NGS. RT-PCR may also be considered if the assay is designed to cover a large number of known fusions [15–20].
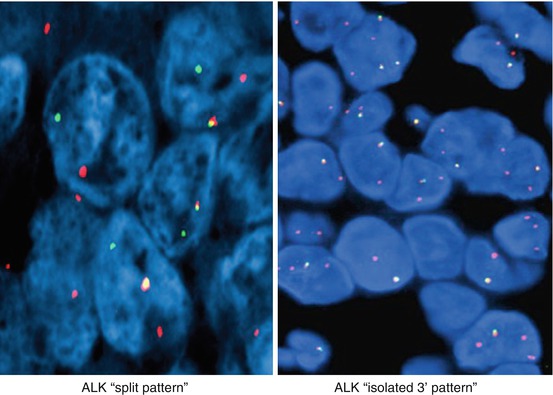

Fig. 8.4
Break-apart ALK FISH translocation patterns
Another major source for false interpretations is the cases in which the rate of rearrangement positive cells falls within the range of 10–20%. In those cases, it is essential to recount the tumor cells and to perform a different assay as indicated above.
ALK false-negative results may also occur and are most likely caused by the complex gene rearrangements and cryptic insertions [14, 21, 22]. Recently, Wiesner et al. identified a novel ALK transcript, ALK ATI, which arises independently of genomic aberrations at the ALK locus through alternative transcription initiation and which can be detected by ALK IHC, but not FISH [23]. Preliminary data showed that the patients with ALK ATI may benefit from ALK inhibitors.
ALK RT-PCR
RT-PCR for detection of ALK rearrangements provides detailed information about ALK fusion partners. The risk of false-negative results and high failure rate for RNA-based assays on FFPE tissue samples make implementation of this assay in clinical practice difficult. In addition, this assay is designed to detect only known fusion partners and fusions with unknown partners would remain undetectable [1, 3, 4, 18, 24, 25].
Other Gene Rearrangements
The other most commonly identified gene fusions identified in lung carcinoma include genes ROS1, RET, NTRK1, and NRG1 . Similar to ALK, break-apart FISH assay is currently the most commonly used method for detection of ROS1 fusions (Fig. 8.5) [26–28]. RET rearrangement is also detectable by FISH or RT-PCR [26, 29–34]. Targeted NGS identified NTRK1 rearrangements resulting in oncogenic fusion products MPRIP-NTRK1 and CD74-NTRK1 [35]. Whole transcriptome sequencing identified CD74-NRG1 fusion that was also identified by RT-PCR [36].
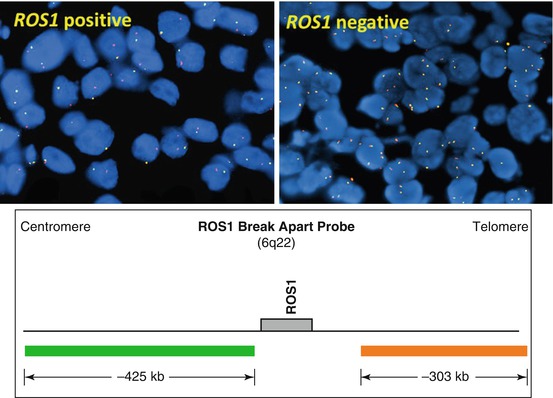

Fig. 8.5
Break-apart ROS1 FISH probe
References
1.
Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med. 2013;137(6):828–60.CrossrefPubMedPubMedCentral
2.
Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–6.CrossrefPubMed
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree