AML, Acute myeloid leukemia; BL, Burkitt’s lymphoma; CLL, chronic lymphocytic leukemia; DLBCL, diffuse large B-cell lymphoma; ES, Ewing sarcoma.
MicroRNAs were found to be involved in the pathophysiology of all types of analyzed human cancers. 18 Among the new paradigms of molecular oncology are the following:
1. Several genome-wide profiling techniques (for review, see References 13, 19, and 20), such as oligonucleotide miRNA microarray, bead-based flow cytometric technique, and quantitative reverse-transcriptase-polymerase chain reaction (qRT-PCR) for precursor and active miRNA or the miRAGE (serial analyses of gene expression for miRNAs), were performed on various cancer histotypes, including chronic lymphocytic leukemia (CLL), breast cancer, glioblastoma, thyroid papillary carcinoma, hepatocellular carcinoma, lung cancer, colon cancer, and endocrine and exocrine pancreatic tumors. From these studies it has become clear that in cancer cells the main alteration of the microRNome (defined as the full complement of microRNAs present in a genome) is represented by aberrant gene expression, consisting of abnormal levels of expression for mature and/or precursor miRNA sequences compared with the corresponding normal tissues.
2. Germline and somatic mutations in miRNAs 21 or polymorphisms in the protein coding mRNAs targeted by miRNAs 22 may also contribute to cancer predisposition, initiation, and progression. In somatic cells, miRNA alterations could initiate or contribute to tumorigenesis, whereas germline mutations could represent cancer-predisposing events.
3. MiRNA profiling achieved by various methods has allowed the identification of signatures associated with diagnosis, staging, progression, prognosis, and response to treatment of human tumors (for review, see Reference 13). Therefore, miRNA “fingerprinting” represents a new addition to the diagnostic and prognostic tools to be used in medical oncology.
Other types of ncRNAs (such as ultraconserved genes [UCGs] and long intergenic noncoding RNAs [lincRNAs]) have also been linked to human cancers. 23 One of the most intriguing characteristics of miRNAs is the near-complete conservation of orthologous genes. For example, the active molecules of the miR-16-1/miR-15a cluster, shown to be an essential player in the initiation of CLL, 21 are completely conserved in humans, mice, and rats and highly conserved in 9 of 10 primate species sequenced. Comparative sequence analysis represents an essential tool in the identification of genomic DNA regions with important biologic functions. Several of these highly conserved genomic sequences were considered not genic (not producing a transcript) and were called conserved nongenic sequences. 24 A special subset of conserved sequences named ultraconserved regions (UCRs) include, by definition, intra- and intergenic sections of the human genome that are absolutely conserved (100% identical with no insertions or deletions) between orthologous regions of the human, rat, and mouse genomes. 25 Because of the high degree of conservation, the UCRs have been demonstrated to have fundamental functional importance for the ontogeny and phylogeny of mammals and other vertebrates. Recently, it was proved that most UCRs are transcribed and that hundreds of UCGs are consistently altered in a significant percentage of analyzed leukemias and carcinomas. UCGs are frequently located at fragile sites and genomic regions involved in cancers. It has also been proven that the inhibition of an overexpressed UCG induces apoptosis in colon cancer cells, and that the expression of some UCGs may be regulated by miRNAs abnormally expressed in CLL. 26 These new regulatory mechanisms support a model in which various types of noncoding genes are actively involved and cooperate with protein-coding genes in human tumorigenesis. Gathering all these notions together makes it clear that noncoding RNA genes, once seen as second-level genomic elements, are now at the center of attention in cancer research. Therefore, it is reasonable to attempt to expand the anticancer ammunition with RNA molecules capable of attenuating or completely abolishing the function of overexpressed ncRNAs—or, alternatively, to reexpress at physiologic levels the deleted or downregulated ncRNAs.
Main Types of Therapeutic RNA Molecules
Three different types of RNA molecules—the ribozymes, the siRNAs, and the anti-miRNA agents—have passed preclinical testing for efficiency in downregulating a target and now are entering clinical trials. At least two more types of RNA molecules recently added to the expanding list of anticancer ammunition—the miRNA-mimic agents and the 8-mer anti-miRNA LNA (locked nucleic acid) molecules—are going through preclinical tests. The antisense oligonucleotide (ASO) strategy was primarily developed using DNA molecules (for detailed discussion, see References 2 and 27). Although the history of RNAs as therapeutic molecules is two decades long, too few clinical trials have yet been conducted on large numbers of cancer patients to allow any convincing conclusion to be drawn. The first hints are encouraging and support the development of new and larger clinical trials. Most of the clinical and preclinical data were gathered from patients with viral infections, such as human immunodeficiency virus type-1 (HIV) or chronic hepatitis C virus (HCV). For a glossary of terms used in RNA inhibition strategies, see Table 55-2 .
Table 55-2
Glossary of Terms in RNA-Inhibition Strategies
Ribozymes
In 1982, the first two ribozymes, the self-splicing intron of the Tetrahymena pre-rRNA and the RNaseP, were discovered by Cech’s group 28 and Altman’s group, 29 respectively. Both shared the Nobel prize in medicine in 1989 for this discovery. A naturally occurring or laboratory-prepared RNA enzyme, ribozyme is an RNA molecule that can catalyze a chemical reaction of substrate cleavage. The mechanism of action consists of three steps that are cyclically repeated. The first is represented by the Watson-Crick base pairing to a complementary target sequence, the second by the site-specific cleavage of the substrate, and the third by the release of the cleavage products. Although there are now seven naturally occurring classes of ribozymes, 30 the most commonly used class of ribozymes as therapeutic agents are the hammerhead (Hh) ribozymes. The Hhs are short-length RNAs, not more than 40 nt long, and are made of two substrate-binding arms plus a conserved catalytic domain of 24 bases. 31
Angiozyme (RPI.4610; Sirna Therapeutics Inc, Boulder, Colo), an angiogenesis inhibitor, is an Hh ribozyme targeting a conserved region of human vascular endothelial growth factor receptor (VEGFR-1), selectively downregulating the VEGFR-1 by cleavage of its mRNA. It is the first synthetic ribozyme to be tested as a therapeutic agent in human cancer. Angiozyme was used in a Phase I clinical trial as a single agent in patients with biopsy-proven refractory solid tumor and showed promising results. The disease was stable in 25% of 28 eligible patients for a period of more than 6 months, with the longest treatment duration of more than 16 months, and showed no significant adverse reactions. 32 In another study, the same drug was combined with carboplatin and paclitaxel, indicating that this multidrug regimen can be administered in patients with advanced solid tumors with no substantial pharmacokinetic interactions. 33 The most common adverse effects were neutropenia, thrombocytopenia, pain, anemia, and fatigue. Angiozyme was well tolerated after intravenous (IV) infusion or a single subcutaneous (SC) bolus in healthy volunteers. 34 Combined with the data from the Phase I clinical trial, new studies will be designed and conducted to assess efficacy in various human cancers. Furthermore, as inhibition of either VEGFR-l or VEGFR-2 signaling can only partially block tumor angiogenesis and growth, simultaneous inhibition of both VEGFR-l and VEGFR-2 signaling could be highly effective in retarding the growth of some tumors. 35
One obvious question is why, in spite of years of effort to decipher the structure and function of ribozymes, data accumulated in cancer patients are so scarce. One reason is the sequence specificity requirements of targeted RNAs that limit the amount of putative targets: for example, the Hh ribozymes work at their best on GUC and AUG triplets. Another reason is the limited accessibility of the drug to the mRNA complementary sequence due to the internal base pairing, producing secondary structures, and to the proteins that physically associate with the RNA. Furthermore, differences in targeted mRNA half-lives could affect the efficacy of the silencing, it being easier to eliminate a target with a shorter half-life than targets with a much longer half-life.
To improve the efficiency of ribozymes in cancer cells, a hybrid construct was produced, referred to as maxizyme. 36 This is a dimer of minimized ribozymes (minizymes) that can cleave two target sites located in two different mRNAs. The maxizyme also can allosterically cleave the target RNA only when it recognizes two target sites. For example, two distinct oncogenes, cyclin Dl (CCND1) and fibroblast growth factor 4 (FGF4, also named HST-1), which are overexpressed in breast cancer cells, were used as targets of a maxizyme. CCND1 activity is required for cell cycle G1/S transition, whereas FGF4 is involved in tumor growth and invasion, and therefore blocking these genes could target a wide spectrum of pathways in malignant cells. When conventional ribozymes were used for suppression of expression of these genes, mRNAs in cancer cells and in normal cells were affected, whereas the trans-maxizyme cleaved these mRNAs only in the breast cancer cells. Whether such a drug can have similar results in clinical trials in patients with breast cancer is still an open question.
siRNAs and shRNAs
In 1998 Mello and Fire discovered RNA interference (RNAi) in vertebrates, 37 for which they received the Nobel prize in medicine in 2006. RNAi is a form of posttranscriptional gene silencing (PTG) in which double-stranded RNA (dsRNA), named siRNA, catalyzes the degradation of complementary mRNA targets. An siRNA is a dsRNA homologous to an mRNA of a target gene. In cytoplasm, the dsRNAs are processed by a complex consisting of Dicer and several other proteins into siRNAs, which are loaded into Argonaute 2 and RNA-induced silencing complex (RISC). The siRNA guide strand recognizes target sites to direct mRNA cleavage, which is carried out by the catalytic domain of AG02. The processing of the siRNAs is similar to that of miRNAs, which is viewed as the “endogenous” process of RNA duplex maturation. 38,39
A small-hairpin RNA (shRNA) represents an siRNA-like molecule expressed from a vector. 40 DNA cassettes encoding RNA polymerase III promoter-driven siRNA-like shRNAs allow long-term expression of therapeutic RNAs in targeted cells. One advantage over the ribozyme technology is the higher efficiency in targeting specific messengers. For example, when the efficiency of the shRNAs was compared with adenoviral delivery of an anti-MDR1 (multidrug resistance) ribozyme construct, the shRNA’s downregulation of mRNA and protein expression was accompanied by a complete inhibition of the pump activity of MDR1 and a reversal of the multidrug-resistant phenotype. The ribozyme construct weakly affects gene expression, confirming that adenoviral delivery of shRNAs is much more effective than adenoviral delivery of ribozymes and that adenovirus-based vectors can be very powerful agents for the efficient delivery of therapeutic RNA molecules. 41
Oncogenes expressed at abnormally high levels represent the main targets of siRNA-directed therapy. Results from preclinical studies are promising and show clear efficacy. For instance, in a mouse model of ovarian cancers overexpressing the tyrosine kinase receptor EphA2 gene, the administration of liposomal-delivered siRNA targeting EphA2 combined with paclitaxel determined a reduction of tumor size greater than 50% with intravenous or intraperitoneal routes of delivery. These data support the idea that chemotherapy and siRNA together can provide a powerful anticancer combination. 42 In another example, adenovirus-mediated siRNA against a K-ras mutated messenger (K-ras codon 12 GGT to GTT) markedly decreased K-ras gene expression and inhibited cellular proliferation of lung cancer cells that express the relevant mutation, but produced only minimal growth inhibition in cells that lack the specific abnormality. 43
Although there are promising preclinical data, siRNA cancer therapy is overshadowed by few, but significant, concerning issues. The first involves the low bioavailability: in aqueous solution, siRNAs are extremely hydrophilic and heavily hydrated because of the exposure of the sugar-phosphate backbone to water, thus reducing their diffusion to the target tissue. Furthermore, because of degradation by serum nucleases, the in vivo half-lives of siRNAs are short. Chemical modifications of siRNAs to overcome these deficiencies include the conformationally “locked” nucleotide analog (LNA), which substantially increases the serum stability, without adversely affecting interactions with cellular silencing machinery. Introduction of a 2′-O-methyl group (2′-O-Me) and 2′-fluoro nucleotides enhances plasma stability and increases potency by several hundredfold in vivo over the unmodified, “naked” siRNAs. 44 Another unsolved problem is represented by off-target effects (OTEs), meaning that, in addition to the complementary target, a specific siRNA can induce the silencing of several other imperfect complementary mRNAs that can be important for cell homeostasis. OTEs have been demonstrated by transcriptional profiling studies, and the nonspecific genes differentially expressed between treated and nontreated cells contain complementary regions to one of the two strands in the siRNA duplex. Chemical modification of nucleotides within this region of homology, by the introduction of a 2′-O-Me group, reduces the OTE without decreasing the silencing activity on messenger RNAs. Finally, a major concern is the stimulation of the innate immune system and the production of high amounts of interferon in response to siRNA duplexes. Sugar modifications (such as 2′-O-Me) and LNAs seem to reduce the immunostimulatory effects of siRNAs (for a comprehensive review of this topic, see Reference 44).
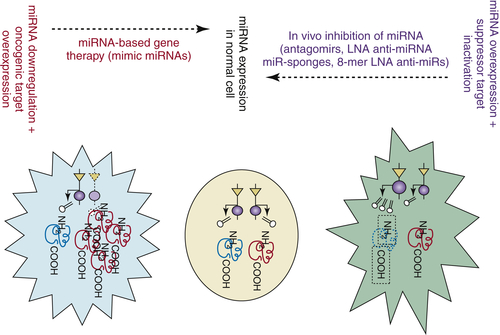
Figure 55-1 MicroRNAs (miRNAs) and anti-miRNAs as new therapeutic agents The type of therapeutic intervention is different with respect to the type of genetic alteration. If overexpressed, agents that specifically reduce miRNA expression to normal levels should be used, whereas if downregulated, agents that restore miRNA expression must be used. (Modified from Pharmacogenomics. 2007;53:521-537.)
ASOs/AMOs Anti-miRNAs, LNAS Anti-miRNAs, and Antagomirs
A rationale for considering miRNAs as potential therapeutic targets is offered by the fact that miRNA overexpression in cancer cells has a pathogenic effect (see preceding sections). Therefore, several types of agents targeting miRNAs are now under development and ready to be tested for their in vivo effects and in clinical trials (Figure 55-1 ). 45,46
Anti-miRNA oligonucleotides (AMOs) represent ASOs, single-stranded, chemically modified DNA-like molecules that are 17 to 22 nt in length and designed to be complementary to a selected miRNA. Thus they are able to specifically inhibit expression of that gene. Mechanistically, AMOs can be described as ASOs against miRNAs and therefore produce an ASO-miRNA duplex through Watson-Crick binding, leading to RNAse-H–mediated cleavage of the target miRNA gene. Important for potential clinical use, ASOs harboring a complete 2′-O-methoxyethyl and phosphorothioate modification have been demonstrated to silence in vivo miR-122 in mouse liver. 47 The LNAs’ anti-miRNAs represent modified antisense single-stranded oligonucleotides 17 to 22 nt in length, with a methylene bridge connecting the 2′ and 4′ carbons. miR-21, shown to be strongly overexpressed in glioblastomas, was silenced in vitro by using LNA-modified antisense oligonucleotides, leading to a significant reduction in cell viability and elevated intracellular levels of caspases. 48
The antagomir represents RNA therapeutic molecules originally designed to inhibit miRNAs. 49,50 These are chemically modified and cholesterol-conjugated single-stranded 23-nt RNA molecules complementary to the targeted miRNAs. The modifications were introduced to increase the stability of the RNA and protect it from degradation. When intravenously administered to mice, antagomirs against miR-122 (antagomir-122), a miRNA highly expressed in liver, induced a marked, specific, and persistent (up to 23 days) reduction of endogenous miRNA gene expression. The same was true for antagomir-16, targeting the ubiquitously expressed miR-16. Silencing of miRNAs by these new agents was followed by physiologic effects, such as a decrease in plasma cholesterol levels after antagomir-122 administration. The only tissue where antagomirs did not act when injected systemically was the brain—probably because of the difficulty of crossing the blood-brain barrier—but they efficiently targeted miRNAs when injected locally into the mouse cortex. One clear advantage with respect to siRNA technology is that antagomirs did not induce an immune response.
A recent addition to this expanding category of therapeutic agents are the 8-mer LNA antimiRs, specifically designed to target the 5′ “seed” region of miRNAs. It was shown that an 8-mer LNA antimiR-155 oligonucleotide targeting miR-155 inhibits Waldenström macroglobulinemia (WM) and CLL cell proliferation in vitro, whereas systemically delivered antimiR-155 showed significantly decreased tumor growth in vivo. 51 Another approach is the use of “miRNA sponges” or “miRNA decoys”—transcripts expressed from strong promoters and containing multiple artificial miRNA binding sites that compete with the endogenous miRNA targets for miRNA binding. 52 Finally, “miRNA masks” are novel gene-specific anti-miRNAs that can selectively inhibit the interaction of the target miRNA with a specific mRNA. These consist of 2′-O-methyl-modified ASOs with locked 3′ and 5′ ends that effectively mask the specific mRNA from the endogenous miRNA and thus prevent its repression. 53
miRNA-Mimic Agents
There are no reports in clinical practice of using the mimic miRNA agents that reproduce the effects of endogenous miRNAs and include all the chemical modifications necessary for stability and processing. However, the restoration of the expression of specific miRNAs abnormally expressed because of genomic deletions, hypermethylation, or other causes could be clinically used in the future. Several studies have validated the efficiency of miRNA mimics in in vitro and in vivo models. For example, intranasal administration of let-7 (which negatively regulates RAS) in a K-ras mutant mouse effectively inhibited tumor growth. 54 Similar studies have been performed for miR-34a in a prostate cancer model, 55 and for miR-26 in a liver cancer model. 56 Thus administration of miRNA-mimetic agents in patients might be a new avenue for clinical cancer management.
In Search of the Right Way and the Right Type of Delivery
Two practical issues regarding the use of RNAs as therapeutic molecules should be further dissected: the selection of the most adequate formula and the most efficient method of administration (local vs. systemic). Naked RNA delivery or vehicle delivered and viral vehicle versus nonviral delivery are the most important questions in the formula type debate. Initial therapeutic applications used 21-nt siRNA duplexes, with 2-nt 3′ overhangs, as well as longer siRNAs of 27 mers and shRNAs (29 nt). These drugs are able to induce only transient gene silencing, because their intracellular concentrations diminish with every cell division. 38 However, despite improvements in siRNA stability, naked siRNAs are of little therapeutic use and are likely to require some form of targeted delivery vehicle for sufficient tissue specificity and cellular uptake. Long-lasting and stable knockdown of target transcription is achieved by expression of shRNAs from various types of vectors, including viral vectors, with Pol II or Pol III promoters. Although therapeutically useful, the continuous expression can cause harmful side effects due to the saturation of the endogenous silencing pathway; thus abundant shRNA expression could be toxic. For example, the use of an adeno-associated virus (AAV) vector with a U6 promoter (Pol III type) induced high expression levels of shRNAs and caused death in about half of treated mice. 57 Different lentiviral vectors (LVs), adenoviral vectors (AVs), and AAVs are being tested. LVs efficiently integrate into the genome of nondividing cells, such as pancreatic islets or terminally differentiated cells. In contrast to LVs, AVs do not integrate into the host genome; they do, however, efficiently transduce dividing and nondividing cells. Different AAV serotypes can be potentially used for organ-directed miRNA expression. Another option is to use nonviral delivery vehicles for shRNAs. Liposomes, lipid complexes with small molecules, polymers, proteins, and antibodies were tested as potential delivery options and all have certain advantages (but also clear disadvantages 58 ). With these delivery partners, the efficacy of shRNA is increased and doses reduced, but the cytotoxic effects are much greater (Table 55-3 ).
A second practical issue is the selection of local versus systemic administration of RNA drugs. Local administration has certain advantages, such as the need for lower doses with consequent lower adverse/toxic reactions and better bioavailability of the drug to the target tissue. Numerous publications have described the delivery of siRNA molecules directly to tumors in vivo, including direct applications of siRNA into to the peritoneal cavity, testes, or urethra (reviewed in Reference 59). In other instances, such as leukemias, local administration cannot be achieved and therefore systemic delivery is mandatory.
A Strategy for Using RNA as Therapeutic Molecules
RNA molecules are now at the center of molecular oncology, with applications for diagnosis and therapy beginning to be proposed. Although exciting and full of promise, the field of RNA as a therapeutic molecule is not free of obstacles. Therefore, a good practical option is to attempt to improve strategies to optimize the efficiency of this new anticancer tool in the light of the unfulfilled promises of other types of new therapies. One approach to cancer therapy is to target various molecular defects in the multistep pathways of specific cancers—the multiplex RNA inhibition targeting strategy. Another strategy is to focus on a major molecular alteration clearly linked to disease pathogenesis by using multiple agents—the Sandwich RNA inhibition strategy. For example, to highlight, let us use the example of B-cell chronic lymphocytic leukemia (B-CLL), the most frequent adult leukemia in the Western world 60 and one of the best studied models of interplay between coding and ncRNAs in human cancers. 21 B-CLL is characterized by predominantly nondividing malignant B cells overexpressing the anti-apoptotic Bc12 protein. Deletion or downregulation of the miR-15a/ miR-16-1 cluster located at chromosome 13q14.3 represents an early event directly involved in the regulation of BCL2 expression. 61 During the evolution of malignant clones, other miRNAs are deleted (such as miR-29) or overexpressed (such as miR-155 or the cluster miR-221/miR-222), contributing to the aggressiveness of B-CLL (Richter syndrome). Such abnormalities influence the expression of other protein-coding genes: reportedly miR-221 and miR-222 directly down-modulate c-KIT receptor and miR-29 regulates the levels of TCL1 oncogene 62 or targets other ncRNAs such as UCGs uc.160 and uc.78 (Figure 55-2 26). The consequences of this steady accumulation of abnormalities are represented initially by low apoptosis and high survival of malignant B cells and later by the evolution of more aggressive clones with a higher proliferative capacity and survival by the overexpression of BCL2 and TCL1 oncogenes. Therefore, a mixed strategy of targeting both these oncogenes at the same time and using combined RNA drugs could be proposed. For example, designing a maxizyme anti-BCL2 and anti-TCL1 could be an option. Combining the systemic delivery of miR-15 and miR-16 family members targeting BCL2, together with miR-29 and miR-181 family members against TCL1, could increase the efficacy of therapeutic oncogenic downregulation. Furthermore, taking advantage of genome-wide profiling technologies, these therapies could be provided to only the subset of CLL patients having both types of lesions, specifically offered only to patients with BCL2 overexpression and mi-R15 and miR-16 downregulations (the indolent form of CLL), or only to patients with TCL1 overexpression and miR-29 and miR-181 downregulation (the aggressive form of CLL).
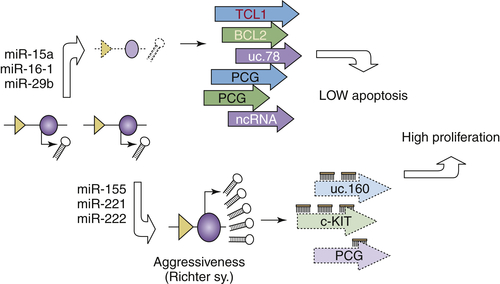
Figure 55-2 Chronic lymphocytic leukemia (CLL) as the best known example of interplay between noncoding RNAs (ncRNAs) and protein coding genes (PCGs) and the therapeutic implications During the initiation and progression of B-cell CLL alteration in at least three different types of genes, PCGs, microRNAs (miRNAs), and ultraconserved genes (UCGs) were identified. These are not independent, as complex regulatory interactions between miRNAs and PCGs and between miRNAs and UCGs occur. Furthermore, we postulate the existence of interactions between UCGs and PCGs, although these have not yet been demonstrated.
Table 55-3
Delivery Methods for RNA-Interference–Based Therapeutics
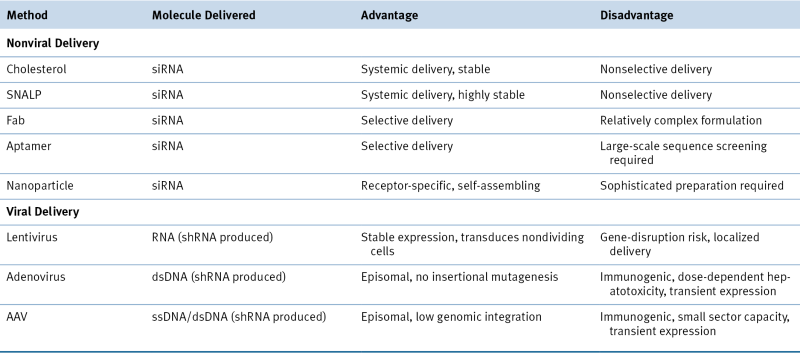
dsDNA, Double-strand DNA; SNALP, stable nucleic acid-lipid particle; ssDNA, single-strand DNA.
Modified with permission from Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173-184.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree