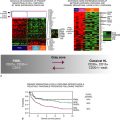
This table is not comprehensive, but provides an example of breast cancer multiplexed gene tests that are currently in development or have been commercialized. The type of sample, number of genes, indication, and ability to predict therapy may change based on commercial development. Prediction of response to therapy has been shown for many of the tests; however, for some of the tests this may not be used or approved as an indication for the test.
Chemo, Chemotherapy; ER, estrogen receptor; FDA, U.S. Food and Drug Administration; FFPE, formalin-fixed paraffin-embedded; IVDMIA, In Vitro Diagnostic Multivariate Index Assay; RT-PCR, reverse transcriptase polymerase chain reaction; TAM, tamoxifen.
Molecular Basis of Breast Cancer
ER Action
Chromatin Remodeling
Growth Factors
Angiogenesis
Conclusion and Outlook
Acknowledgments
1. http://planning.cancer.gov/library/1998breastcancer.pdf .
2. Modelling breast cancer: one size does not fit all . Nat Rev Cancer . 2007 ; 7 : 659 – 672 .
3. Hormone action in the mammary gland . Cold Spring Harb Perspect Biol . 2010 ; 2 a003178 .
4. Growth factor signalling in endocrine and anti-growth factor resistant breast cancer . Rev Endocr Metab Disord . 2007 ; 8 : 241 – 253 .
5. Generation of a functional mammary gland from a single stem cell . Nature . 2006 ; 439 : 84 – 88 .
6. Purification and unique properties of mammary epithelial stem cells . Nature . 2006 ; 439 : 993 – 997 .
7. Control of mammary stem cell function by steroid hormone signalling . Nature . 2010 ; 465 : 798 – 802 .
8. Progesterone induces adult mammary stem cell expansion . Nature . 2010 ; 465 : 803 – 807 .
9. BRCA1 regulates human mammary stem/progenitor cell fate . Proc Natl Acad Sci U S A . 2008 ; 105 : 1680 – 1685 .
10. The cell of origin of BRCA1 mutation-associated breast cancer: a cautionary tale of gene expression profiling . J Mammary Gland Biol Neoplasia . 2011 ; 16 : 51 – 55 .
11. The clonal evolution of tumor cell populations . Science . 1976 ; 194 : 23 – 28 .
12. Stem cells, cancer, and cancer stem cells . Nature . 2001 ; 414 : 105 – 111 .
13. Cells of origin in cancer . Nature . 2011 ; 469 : 314 – 322 .
14. Tumor heterogeneity: causes and consequences . Biochim Biophys Acta . 2010 ; 1805 : 105 – 117 .
15. Ductal carcinoma in situ: terminology, classification, and natural history . J Natl Cancer Inst Monogr . 2010 ; 41 : 134 – 138 .
16. Analysis of loss of heterozygosity in 399 premalignant breast lesions at 15 genetic loci . J Natl Cancer Inst . 1998 ; 90 : 697 – 703 .
17. Molecular diversity in ductal carcinoma in situ (DCIS) and early invasive breast cancer . Mol Oncol . 2010 ; 4 : 357 – 368 .
18. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution . Clin Cancer Res . 2008 ; 14 : 370 – 378 .
19. The molecular pathology of breast cancer progression . J Pathol . 2011 ; 223 : 307 – 317 .
20. Clinical management of hereditary breast cancer syndromes . J Mammary Gland Biol Neoplasia . 2011 ; 16 : 17 – 25 .
21. Translational advances regarding hereditary breast cancer syndromes . J Surg Oncol . 2012 ; 105 : 444 – 451 .
22. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing . Proc Natl Acad Sci U S A . 2010 ; 107 : 12629 – 12633 .
23. Ten genes for inherited breast cancer . Cancer Cell . 2007 ; 11 : 103 – 105 .
24. Mutation of a single allele of the cancer susceptibility gene BRCA1 leads to genomic instability in human breast epithelial cells . Proc Natl Acad Sci U S A . 2011 ; 108 : 17773 – 17778 .
25. Genetic interactions in cancer progression and treatment . Cell . 2011 ; 145 : 30 – 38 .
26. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers . N Engl J Med . 2009 ; 361 : 123 – 134 .
27. PARP inhibitors in breast cancer: BRCA and beyond . Oncology (Williston Park) . 2011 ; 25 : 1014 – 1025 .
28. Reproductive Factors . New York, NY : Springer ; 2010 : 119–137 .
29. An update on ovarian suppression/ablation . Int J Gynecol Cancer . 2006 ; 16 ( suppl 2 ) : 511 – 514 .
30. Hormone replacement therapy and the risk of breast cancer . Nat Rev Clin Oncol . 2011 ; 8 : 669 – 676 .
31. Aspects of the rationale for the Women’s Health Trial . J Natl Cancer Inst . 1988 ; 80 : 802 – 814 .
32. The Women’s Health Initiative Randomized Controlled Dietary Modification Trial: a post-mortem . Breast Cancer Res Treat . 2009 ; 114 : 1 – 6 .
34. Traditional breast cancer risk factors in relation to molecular subtypes of breast cancer . Breast Cancer Res Treat . 2012 ; 131 : 159 – 167 .
35. Breast cancer prognostic classification in the molecular era: the role of histological grade . Breast Cancer Res . 2010 ; 12 : 207 .
36. Steroid receptors in human breast cancer . Cancer Res . 1978 ; 38 : 4289 – 4291 .
37. Biology of progesterone receptor loss in breast cancer and its implications for endocrine therapy . J Clin Oncol . 2005 ; 23 : 7721 – 7735 .
38. Treatment of HER2-positive breast cancer: current status and future perspectives . Nat Rev Clin Oncol . 2012 ; 9 : 16 – 32 .
39. Current treatment options in triple negative breast cancer . Breast Dis . 2010 ; 32 : 99 – 122 .
40. Practical implications of gene-expression-based assays for breast oncologists . Nat Rev Clin Oncol . 2012 ; 9 : 48 – 57 .
41. Molecular portraits of human breast tumours . Nature . 2000 ; 406 : 747 – 752 .
42. Repeated observation of breast tumor subtypes in independent gene expression data sets . Proc Natl Acad Sci U S A . 2003 ; 100 : 8418 – 8423 .
43. Show me the data! . Nat Genet . 2001 ; 29 : 373 .
44. Molecular classification of estrogen receptor-positive/luminal breast cancers . Adv Anat Pathol . 2012 ; 19 : 39 – 53 .
45. Luminal-B breast cancer and novel therapeutic targets . Breast Cancer Res . 2011 ; 13 : 221 .
46. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups . Nature . 2012 ; 486 : 346 – 352 .
47. Basal cytokeratins and their relationship to the cellular origin and functional classification of breast cancer . Breast Cancer Res . 2005 ; 7 : 143 – 148 .
48. Breast cancer molecular profiling with single sample predictors: a retrospective analysis . Lancet Oncol . 2010 ; 11 : 339 – 349 .
49. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes . Cancer Cell . 2006 ; 10 : 515 – 527 .
50. Single-cell dissection of transcriptional heterogeneity in human colon tumors . Nat Biotechnol . 2011 ; 29 : 1120 – 1127 .
51. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing . Genome Res . 2011 ; 21 : 741 – 747 .
52. Identification of allele-specific alternative mRNA processing via transcriptome sequencing . Nucleic Acids Res . 2012 ; 40 : e104 .
53. Noncoding RNAs involved in mammary gland development and tumorigenesis: there’s a long way to go . J Mammary Gland Biol Neoplasia . 2012 ; 17 : 43 – 58 .
54. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer . Nat Med . 2011 ; 17 : 1646 – 1651 .
55. Characterization of breast cancer by array comparative genomic hybridization . Biochem Cell Biol . 2007 ; 85 : 497 – 508 .
56. Breast cancer genomes—form and function . Curr Opin Genet Dev . 2010 ; 20 : 4 – 14 .
57. Novel patterns of genome rearrangement and their association with survival in breast cancer . Genome Res . 2006 ; 16 : 1465 – 1479 .
58. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses . Genome Res . 2009 ; 19 : 521 – 532 .
59. A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome . Genome Res . 2009 ; 19 : 167 – 177 .
60. Complex landscapes of somatic rearrangement in human breast cancer genomes . Nature . 2009 ; 462 : 1005 – 1010 .
61. Massive genomic rearrangement acquired in a single catastrophic event during cancer development . Cell . 2011 ; 144 : 27 – 40 .
62. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements . Cell . 2011 ; 146 : 889 – 903 .
63. Whole-genome analysis informs breast cancer response to aromatase inhibition . Nature . 2012 ; 486 : 353 – 360 .
64. The landscape of cancer genes and mutational processes in breast cancer . Nature . 2012 ; 486 : 400 – 404 .
65. The clonal and mutational evolution spectrum of primary triple-negative breast cancers . Nature . 2012 ; 486 : 395 – 399 .
66. Sequence analysis of mutations and translocations across breast cancer subtypes . Nature . 2012 ; 486 : 405 – 409 .
67. Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways . Cancer Treat Rev . 2012 ; 38 : 698 – 707 .
68. Gene-expression-based prognostic assays for breast cancer . Nat Rev Clin Oncol . 2010 ; 7 : 340 – 347 .
69. Individualization of therapy using Mammaprint: from development to the MINDACT Trial . Cancer Genomics Proteomics . 2007 ; 4 : 147 – 155 .
70. The predictive value of the 70-gene signature for adjuvant chemotherapy in early breast cancer . Breast Cancer Res Treat . 2010 ; 120 : 655 – 661 .
71. Prognostic utility of the breast cancer index and comparison to Adjuvant! Online in a clinical case series of early breast cancer . Breast Cancer Res . 2011 ; 13 : R98 .
72. A 50 gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen . Clin Cancer Res . 2012 ; 18 : 4465 – 4472 .
73. Gene expression profiling in breast cancer: classification, prognostication, and prediction . Lancet . 2011 ; 378 : 1812 – 1823 .
74. Estrogen receptors: how do they signal and what are their targets . Physiol Rev . 2007 ; 87 : 905 – 931 .
75. Mechanisms of endocrine resistance in breast cancer . Annu Rev Med . 2011 ; 62 : 233 – 247 .
76. An oestrogen-receptor-alpha-bound human chromatin interactome . Nature . 2009 ; 462 : 58 – 64 .
77. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells . Cell . 2011 ; 145 : 622 – 634 .
78. Steroid receptor coactivators 1, 2, and 3: critical regulators of nuclear receptor activity and steroid receptor modulator (SRM)-based cancer therapy . Mol Cell Endocrinol . 2012 ; 348 : 430 – 439 .
79. Inverse relationship between ER-beta and SRC-1 predicts outcome in endocrine-resistant breast cancer . Br J Cancer . 2004 ; 91 : 1687 – 1693 .
80. Expression of SRC-1, AIB1, and PEA3 in HER2 mediated endocrine resistant breast cancer; a predictive role for SRC-1 . J Clin Pathol . 2004 ; 57 : 1069 – 1074 .
81. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer . J Natl Cancer Inst . 2003 ; 95 : 353 – 361 .
82. The p160 ER co-regulators predict outcome in ER negative breast cancer . Breast Cancer Res Treat . 2012 ; 131 : 463 – 472 .
83. Expression analysis of estrogen receptor alpha coregulators in breast carcinoma: evidence that NCOR1 expression is predictive of the response to tamoxifen . Clin Cancer Res . 2003 ; 9 : 1259 – 1266 .
84. Minireview: Recent advances in extranuclear steroid receptor actions . Endocrinology . 2011 ; 152 : 4489 – 4495 .
85. International Journal of Breast Cancer, vol. 2011, Article ID 232435, 10 pages, 2011. doi:10.4061/2011/232435
86. Cytoplasmic estrogen receptor in breast cancer . Clin Cancer Res . 2012 ; 18 : 118 – 126 .
87. Growth factor stimulation induces a distinct ER(alpha) cistrome underlying breast cancer endocrine resistance . Genes Dev . 2010 ; 24 : 2219 – 2227 .
88. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer . Nature . 2012 ; 481 : 389 – 393 .
89. Tamoxifen treatment for breast cancer: concept to gold standard . Oncology (Williston Park) . 1997 ; 11 : 7 – 13 .
90. Development and evolution of therapies targeted to the estrogen receptor for the treatment and prevention of breast cancer . Steroids . 2007 ; 72 : 7 – 25 .
91. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen . Cell . 1998 ; 95 : 927 – 937 .
92. A pharmacological review of selective oestrogen receptor modulators . Hum Reprod Update . 2000 ; 6 : 212 – 224 .
93. Aromatase, aromatase inhibitors, and breast cancer . J Steroid Biochem Mol Biol . 2011 ; 125 : 13 – 22 .
94. Mechanisms of estrogen-independent breast cancer growth driven by low estrogen concentrations are unique versus complete estrogen deprivation . Breast Cancer Res Treat . 2012 ; 134 : 1027 – 1039 .
95. The androgen metabolite 5alpha-androstane-3beta,17beta-diol (3betaAdiol) induces breast cancer growth via estrogen receptor: implications for aromatase inhibitor resistance . Breast Cancer Res Treat . 2009 ; 115 : 289 – 296 .
96. Role of fulvestrant in the management of postmenopausal breast cancer . Expert Rev Anticancer Ther . 2011 ; 11 : 1641 – 1652 .
97. Activity of fulvestrant 500 mg versus anastrozole 1 mg as first-line treatment for advanced breast cancer: results from the FIRST study . J Clin Oncol . 2009 ; 27 : 4530 – 4535 .
98. Combination anastrozole and fulvestrant in metastatic breast cancer . N Engl J Med . 2012 ; 367 : 435 – 444 .
99. Epigenetics as a therapeutic target in breast cancer . J Mammary Gland Biol Neoplasia . 2012 ; 17 : 191 – 204 .
100. Epigenetics in breast cancer: what’s new? Breast Cancer Res . 2011 ; 13 : 225 .
101. Somatic mutations of the mixed-lineage leukemia 3 (MLL3) gene in primary breast cancers . Pathol Oncol Res . 2011 ; 17 : 429 – 433 .
102. EZH2 methyltransferase and H3K27 methylation in breast cancer . Int J Biol Sci . 2012 ; 8 : 59 – 65 .
103. Growth factors in breast cancer . Endocr Rev . 1995 ; 16 : 559 – 589 .
104. HER2: biology, detection, and clinical implications . Arch Pathol Lab Med . 2011 ; 135 : 55 – 62 .
105. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2 . N Engl J Med . 2001 ; 344 : 783 – 792 .
106. Trastuzumab in the adjuvant treatment of HER2-positive early breast cancer patients: a meta-analysis of published randomized controlled trials . PLoS One . 2011 ; 6 : e21030 .
107. EMILIA Study Group. Trastuzumab emtansine for HER2-positive advanced breast cancer . N Engl J Med . 2012 ; 367 ( 19 ) : 1783 – 1791 .
108. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer . N Engl J Med . 2012 ; 366 : 109 – 119 .
109. ErbB family receptor inhibitors as therapeutic agents in breast cancer: current status and future clinical perspective . Med Res Rev . 2012 ; 32 : 166 – 215 .
110. Disruption of steroid and prolactin receptor patterning in the mammary gland correlates with a block in lobuloalveolar development . Mol Endocrinol . 2002 ; 16 : 2675 – 2691 .
111. Growth hormone and insulin-like growth factor-I in the transition from normal mammary development to preneoplastic mammary lesions . Endocr Rev . 2009 ; 30 : 51 – 74 .
112. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen . Nature . 2008 ; 456 : 663 – 666 .
113. Cross talk between estrogen receptor and IGF signaling in normal mammary gland development and breast cancer . Breast Dis . 2003 ; 17 : 105 – 114 .
114. High IGF-IR activity in triple-negative breast cancer cell lines and tumorgrafts correlates with sensitivity to anti-IGF-IR therapy . Clin Cancer Res . 2011 ; 17 : 2314 – 2327 .
115. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer . N Engl J Med . 2012 ; 366 : 520 – 529 .
116. Tumorigenesis and the angiogenic switch . Nat Rev Cancer . 2003 ; 3 : 401 – 410 .
117. Controlling angiogenesis in breast cancer: a systematic review of anti-angiogenic trials . Cancer Treat Rev . 2012 ; 38 : 673 – 688 .
118. Modes of resistance to anti-angiogenic therapy . Nat Rev Cancer . 2008 ; 8 : 592 – 603 .
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree