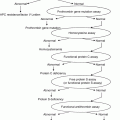
CYP2C9
VKORC1
*1/*1
*1/*2
*1/*3
*2/*2
*2/*3
*3/*3
GG
Typical
Typical
Moderate ↓
Moderate ↓
Moderate ↓
Significant ↓
AG
Typical
Moderate ↓
Moderate ↓
Moderate ↓
Significant ↓
Significant ↓
AA
Moderate ↓
Moderate ↓
Significant ↓
Significant ↓
Significant ↓
Significant ↓
Several algorithms are available that utilize both genotype and clinical information to predict warfarin maintenance dose requirements [1, 8, 9]. One such algorithm is available at www.WarfarinDosing.org. This algorithm takes into account numerous clinical factors, including certain concomitant medications, and allows the information to be saved and modified based on additional information from INR monitoring. In a study of this algorithm, the pharmacogenetic equation used explained a little over 50 % of the variability in warfarin dose in both derivation (n = 1015) and validation (n = 292) cohorts [8]. Performance of this algorithm was superior to equations that accounted only for clinical factors, which explained approximately 20 % of dose variability [8]. Current algorithms are limited by a lack of detailed guidance regarding dosing intervals and laboratory monitoring.
Small prospective studies have compared the use of pharmacogenetics-guided warfarin dosing with traditional management. These studies have shown mixed results, with three showing no difference in the time in therapeutic range (TTR) and one showing increased TTR in the pharmacogenetic-guided group [1, 10–12]. These small studies did not show differences in thrombotic events, major bleeding, or survival [10]. In 2013, results from three randomized controlled trials were reported in the New England Journal of Medicine. Two of these trials compared genotype-guided dose determination with clinical algorithms and showed no significant differences in the primary outcome of TTR or other endpoints [13, 14]. The third trial compared a pharmacogenetic algorithm with the local standard of care [15]. In this study, genotype-guided dosing was associated with a higher percentage of TTR than standard dosing during warfarin initiation.
When estimating warfarin dosing, it is critically important to consider potential interactions with other medications, including drugs with possible synergistic anticoagulant effects, impact on vitamin K status, or effects on warfarin metabolism. Caution must be used not only with prescription medications, but also over-the-counter and herbal preparations. Numerous medications are metabolized by the liver cytochrome P450 system and may either inhibit or induce these enzymes, with potential impacts on warfarin metabolism. Common drugs that may influence required warfarin does include amiodarone, statins, antibiotics, and antifungals.
There are several FDA-cleared warfarin genotyping assays utilizing a variety of methodologies [5]. These tests identify the CYP2C9*2 and *3 alleles and the VKORC1 promoter mutation −1639G > A, but do not detect other warfarin sensitivity variants or mutations causing warfarin resistance [5]. The wild-type allele (*1) is not directly detected, but is inferred when other variants are not identified, a situation that can lead to genotyping errors in the presence of untested polymorphisms [5, 6]. In an analysis of 5 years of proficiency testing (PT) data from the College of American Pathologists (CAP), participating laboratories demonstrated greater than 98 % accuracy in identifying samples with the wild type CYP2C9 diplotype (*1/*1, 10 surveys), as well as samples containing variant alleles with diplotypes of *1/*2 (4 surveys), *1/*3 (4 surveys), and *2/*3 (2 surveys) [6]. Similarly, accuracy in identifying wild-type (G/G) versus variant (GA and AA) genotypes of VKORC1 exceeded 96 % in ten total surveys [6]. In the CAP PT analysis, laboratories were also asked to provide expected phenotype. Genotype-phenotype concordance ranged from 89 to 96 % for CYP2C9, with the lowest values seen for prediction of the *1/*2 and *1/*3 phenotypes, and from 89 to 94 % for VKORC1, with the lowest value for prediction of the AA phenotype [6]. Laboratories providing genotyping results should provide guidance on expected phenotype, but are generally limited in their ability to provide comprehensive information due to the need for detailed clinical data. Although current tests can yield results in a few hours, testing availability is usually limited to reference laboratories or larger hospital-based laboratories, which may limit utility to impact initial therapy [16]. Point-of-care assays for warfarin genotyping have been studied, but are not widely used [17]. Problems with reimbursement of warfarin pharmacogenetic testing are an ongoing issue for clinical services and laboratories. This testing is not currently reimbursed by Medicare outside of clinical trial settings [5].
Despite the theoretical advantages of warfarin dose adjustment based on genotyping, the testing is not currently considered the standard of care. Current practice guidelines from the American College of Chest Physicians and American College of Medical Genetics do not recommend routine use of pharmacogenetic testing to guide warfarin dosing at the initiation of therapy due to a lack of evidence supporting this approach and probable lack of cost-effectiveness [3, 10]. There are specific groups where pharmacogenetic testing may be useful including medication-compliant subjects requiring unusually low or high doses to maintain a therapeutic INR, or subjects who may be prescribed prophylactic warfarin in the future [3]. Pharmacogenetic-guided warfarin dosing, if utilized, does not eliminate the need for close clinical follow-up and laboratory monitoring via the INR.
11.3 Pharmacogenetic Testing for Clopidogrel
Clopidogrel is an oral thienopyridine antiplatelet medication that irreversibly blocks the platelet surface P2Y12 ADP receptor, inhibiting platelet aggregation and reducing the risk of adverse cardiovascular outcomes in patients with cardiovascular disease [18]. Clopidogrel is administered as an inactive prodrug that undergoes a 2-step conversion to an active thiol metabolite by the liver cytochrome P450 (CYP) system [19]. Several CYP enzymes, including CYP2C19, CYP3A, and CYP2B6, and CYP1A2, are involved in the metabolism of clopidogrel, but CYP2C19 is considered the principle enzyme responsible for its activation [18–20]. Genetic polymorphisms in CYP2C19 influence generation of the active metabolite. In March 2010, after several previous labeling changes, the Food and Drug Administration (FDA) added a boxed warning to the package insert of clopidogrel informing prescribing physicians about potential risks for adverse cardiovascular events in clopidogrel poor metabolizers who generate lesser amounts of the active metabolite [19]. The warning states that testing is available to identify a patient’s CYP2C19 genotype and advises consideration of alternative treatment or treatment strategies in subjects who are poor metabolizers. Alternative treatment strategies could include administering higher doses, replacing clopidogrel with other agents such as prasugrel or ticagrelor, or addition of a third antiplatelet agent to dual antiplatelet regimens, although evidence-based alternatives have not been established in clinical trials [19]. Prasugrel is a more potent P2Y12 inhibitor that is converted to its active metabolite in a single step and has very few reports of poor responders [19, 21, 22]. However, prasugrel is associated with increased rates of bleeding as compared to clopidogrel [19]. Ticagrelor is a reversible non-thienopyridine P2Y12 inhibitor that does not require activation [19, 23].
The CYP2C19*1 allele is the wild-type allele (full function) and several alleles associated with reduced or absent function have been described (*2, *3, *4, *5, *6, *7, and *8, others). CYP2C19*2 and *3 are non-functional and represent the most common reduced function alleles in whites and Asians [19]. The *2 allele (24 % of whites, 30 % of African Americans, and 50 % of Asians have at least one copy) encodes a cryptic splice site that results in complete loss of enzyme activity, while the *3 allele results in introduction of a stop codon [20, 24]. Using the CYP2C19 genotype, individuals can be phenotypically classified as ultra-rapid, extensive, intermediate, or poor metabolizers (see Table 11.2) [6, 25]. Extensive metabolizers have no loss-of-function (LOF) alleles and generate expected amounts of active drug, intermediate metabolizers carry 1 LOF allele, and poor metabolizers carry 2 LOF alleles (2 to 14 % of the population) [25, 26]. Several studies have revealed an increased risk of major adverse cardiac events in subjects with reduced function alleles, particularly poor metabolizers who have significantly lower exposure to the active metabolite and less inhibition of platelet aggregation [19, 26–28].
Table 11.2
Summary of CYP2C19 variants and effect on clopidogrel metabolism
CYP2C19 variant | Description | Clopidogrel metabolism status | Possible therapeutic approacha |
|---|---|---|---|
*1 | Wild-type | *1/*1 = extensive metabolizer | Standard clopidogrel dosing |
*2 or *3 | Loss of function | 1 allele = intermediate metabolizer | Consider alternative antiplatelet therapy if not contraindicated |
2 alleles = poor metabolizer | |||
*17 | Increased function | *1/*17 or *17*17 = ultra-rapid metabolizerb | Standard clopidogrel dosing |
Clinically available CYP2C19 genotyping tests generally characterize the most common LOF alleles CYP2C19*2 and *3 by targeted real-time polymerase chain reaction (PCR). Some tests also evaluate less common LOF alleles and may look for *17, which is associated with ultra-rapid metabolism, increased exposure to the active metabolite, increased platelet inhibition, and increased risk of bleeding (see Table 11.2) [26]. The *17 variant (3–21 % of the population) results from a promoter mutation that results in increased CYP2C19 gene transcription and this hyper-functional variant interacts with LOF alleles, if present [20, 25]. As in the warfarin genotyping tests, the presence of the wild-type allele (*1) is often inferred when other variants are not identified, a situation that can result in systematic genotyping errors when there are variants present that are not identified by the testing panel [6]. When interpreting results, it is important to know which CYP2C19 variants can and cannot be identified by the test.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree