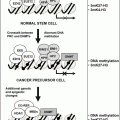
Fig. 8.1
Critical signaling pathways as drug targets for cancer therapy. (a) Surface receptors dimerize and activate downstream effectors such as RAS (a GTPase). Many surface receptors are targeted by antibodies, and EGFR has been targeted through cetuximab and trastuzumab. (b) RAS activation is caused by SOS-mediated exchange of GDP for GTP on RAS. G protein signaling molecules, such as RAS, have been targeted by farnesyltransferase inhibitors (FTI) and S-farnesylthiosalicylic acid (FTS). (c) The BCR-ABL fusion protein activates RAS, PI3K, and other oncogenic signaling molecules. Tyrosine kinases are often targeted with small molecules. BCR-ABL has been targeted by Imanitib. (d) Transcription factors are often the terminal effectors of a pathway. To date, transcription factors are yet to be successfully targeted. All drugs are in red at their sites of action.
8.2.1 Receptor Signaling
Cell surface receptors are the vehicles of communication amongst cells. They are responsible for initiating the response of cells to their tissue microenvironment and mediate many signaling cascades. Growth factor receptors were some of the first proto-oncogenes discovered [2]. A multitude of cell surface receptors have been implicated in tumorigenesis including the epithelial growth factor receptor (EGFR), platelet-derived growth factor receptor (PDGFR), and insulin-like growth factor receptor (IGFR) [3]. Each of these receptors has been implicated in the pathogenesis of specific types of human neoplasia.
In many regards the ErbB family of transmembrane tyrosine kinase receptors is prototypical of cell surface receptors. This gene family includes four members: (1) EGFR (ErbB1), (2) HER2/NEU (ErbB2 hereafter referred to as HER2), (3) ErbB3, and (4) ErbB4. Ligands have been identified for each of the ErbB family members with the notable exception of HER2 that likely heterodimerizes with EGFR or HER3. Downstream effectors of these molecules include many other proto-oncogenes including the MAPK/RAF/RAS, PI3K/AKT, and STAT signaling. Activation of these receptor pathways induces cellular proliferation, enhanced survival, and increased cellular motility [4].
Many of these gene products have multiple alleles that appear to exhibit tissue-specific expression. For example, each of the ErbB receptors exhibits unique expression patterns. Correspondingly, mutated receptors have been implicated in particular types of human cancer [5]. For example, EGFR is commonly overexpressed in lung carcinoma [6], while Her2 overexpression is more commonly associated with breast cancer [7]. Thus, the biology and pathophysiology associated with these gene products is associated with specific cellular lineages.
Cell surface receptors are particularly attractive candidates as drug targets. First, receptors are specific to particular tissue types thus allowing for target specificity. Second, pharmacologically blocking a ligand-binding site is an obvious and frequently successful strategy to inactivate a receptor. Finally, drugs that target receptor molecules do not need to be able to transit through cellular membranes.
8.2.2 GTPases
GTPases are a large family of gene products that mediate receptor signaling [8]. A multitude of GTPases have been implicated as oncogenes. A characteristic feature of the biology of GTPases is that they only become activated after posttranslational modification that permits their localization to the plasma membrane [9]. Moreover, the enzymes that mediate these modifications are possible targets for the treatment of cancer.
The RAS family represents a prototypical example of GTPases. There are three members: (1) H-RAS, (2) K-RAS, and (3) N-RAS [8]. A mutant RAS was one of the first oncogenes identified to be mutated in a human cancer. Over 25 % of human cancers exhibit activating point mutations of a RAS gene [10]. RAS proteins are known to play a critical role in the regulation of a multitude of cellular programs including cellular growth, proliferation, apoptosis, and angiogenesis [9, 11]. The RAS/RAF/MAPK pathway is a frequent target for pharmacologic treatment of human cancers.
8.2.3 Protein Kinases
Protein Kinases are among the most abundant signaling molecules with approximately 500 members. The phosphorylation of proteins at serine, threonine, or tyrosine represents one of the most common and fundamental mechanisms of cellular signaling. Phosphorylation has been implicated in the regulation of the activity of cellular receptors, signaling molecules, transcription factors, DNA repair molecules, and chromatin structure proteins amongst many different functions. Many of these gene products when mutated exhibit oncogenic function through constitutive activation [12]. The recent ability to pharmacologically target the ATP binding domains of kinases has made these gene products one of the most frequently pharmacologically targeted molecules [13, 14].
An important protein kinase associated with neoplasia is the BCR-ABL fusion protein. BCR-ABL results from a chromosomal translocation between the ABL proto-oncogene and the BCR locus. BCR-ABL overexpression has been implicated in the pathogenesis of both chronic myelogenous leukemia and acute lymphocytic leukemia. Targeted inactivation of BCR-ABL through the drug imatanib mesylate, is the most regarded example of a successful targeted therapeutic (Gleevec, STI-571) [15–18].
8.2.4 Transcription Factors
Transcription factors are some of the most potent oncoproteins in the induction of cancer. By regulating the expression of numerous gene products they can exert broad, concerted, and sustained effects on cellular programming. They include a multitude of gene products including steroid receptors and nuclear transcription factors such as MYC, MYB, and FOS/JUN.
The family of MYC proto-oncogenes (c-MYC, n-MYC, l-MYC) have been implicated in the regulation of many physiologic programs cellular growth, proliferation, differentiation, immortalization/senescence, apoptosis, angiogenesis, adhesion, motility, and DNA repair. MYC is overexpressed in over half of all human cancers suggesting that it plays a role in the pathogenesis of most human cancers [19]. MYC has been shown to regulate the transcription of up to thousands of target genes [20–22], suggesting that these gene products function as grand coordinators of gene expression programs. However, MYC has also been shown to play a role in the regulation of ribosomal biogenesis, RNA processing, DNA replication, and DNA repair.
c-MYC was one of the first proto-oncogenes identified [23]. c-MYC is activated in human cancer through at least three different mechanisms. The chromosomal translocation of the MYC locus to the immunoglobulin or T-cell receptor loci is associated with Burkitt’s lymphoma. n-MYC is commonly expressed in neuronal cells and is often overexpressed as a result of genomic amplification in neuroblastoma. l-MYC was first identified in lung tissue and is associated with small cell lung carcinoma [19]. MYC has also shown to be overexpressed as a consequence of the disruption of the regulation of its transcription regulations, for example, through the disruption of the beta-catenin pathway through mutation of the APC gene. Although MYC and other transcription factors play such a fundamental role in the pathogenesis of cancer, they are yet to be successfully targeted therapeutically.
8.3 Interrogating Oncogenes via Mouse Models
Our understanding of the physiologic function of proto-oncogenes and the contribution of oncogenes to the initiation and maintenance of tumorigenesis has been markedly enhanced by the ability to generate mouse models. Most notably, the development of transgenic mouse models has provided the unprecedented opportunity to model the contribution of specific gene products to the pathogenesis of neoplasia in vivo. The use of conditional transgenic models has made it possible to directly interrogate when and how the inactivation of oncogenes can result in cancer regression.
Several recent reviews have extensively described the application of conventional transgenic mouse models for the development of therapeutics for cancer [24–26]. Here we will focus on the use of conditional transgenic models can be used to define when and how oncoproteins can be used as targets for the treatment of cancer.
8.3.1 Experimental Approaches
Three different strategies have been most commonly utilized to conditionally regulate gene expression in transgenic mouse models: (1) the Tet System, (2) the Tamoxifen System, and (3) the TVA system [24, 27, 28].
8.3.1.1 The Tet System
The Tetracycline Regulatory System (Tet System ) was developed as a strategy to regulate the transcription of genes in eukaryotic cells utilizing prokaryotic transcriptional regulatory proteins [29, 30]. There are two variations of this system—one activates transgene expression in the presence of a tetracycline such as doxycycline (Tet-on), while the other system shuts off transgene expression upon doxycycline addition (Tet-off). In both variations, two different transgenes are generated. The first transgene uses a tissue-specific promoter to drive the expression of a tetracycline transactivator (tTA or rtTA). The second transgene contains a tetracycline response element (Tet-O) adjacent to a target gene of interest. The tTA or rtTA protein binds to the Tet-O promoter regulating gene transcription. The presence of doxycycline, prevents binding of the tTA protein to the Tet-O element turning off gene expression (Tet-off) or promotes the binding of the rtTA protein to the Tet-O element turning gene expression on (Tet-on). The Tet system facilitates monitoring of transgene expression at the transcription level in specific tissues within the mouse.
8.3.1.2 The Tamoxifen System
The Tamoxifen System also has been employed to conditionally regulate gene activation posttranscriptionally. MYC fused with the estradiol receptor exhibited conditional oncogene activation [31]. A mutant version of the estradiol receptor, which binds tamoxifen, is utilized to prevent endogenous estradiol from activating gene function [32]. Upon addition of tamoxifen, MYC is active, and withdrawal leads to an inactive product. Subsequently, this strategy was shown to be useful in the generation of conditional transgenic mice [33, 34].
8.3.1.3 The RCAS-TVA-Tet System
The Tet-system can be combined with the RCAS-TVA system [35, 36]. In this approach a tissue-specific promoter is used to drive the expression of the avian retroviral receptor (TVA) in transgenic mice. The cells of these mice also contain a Tet-O regulated transgene, but lack the rtTA protein. Avian retroviral vectors (RCAS) are used to deliver the rtTA transactivator to cells that express the TVA. The successfully infected cells now contain a transgene whose expression can be regulated by doxycycline.
8.3.2 Defining When Cancer Is Reversible
Conditional transgenic models have been used to evaluate the consequences of oncogene inactivation in vivo. From these studies, several general themes emerge regarding the role of oncogenes in the initiation and maintenance of tumorigenesis, as we have described [27, 37–42]. Oncogene inactivation can reverse tumorigenesis by inducing sustained cancer regression through differentiation, proliferative arrest, and/or apoptosis (Table 8.1).
Table 8.1
Consequences of oncogene inactivation in transgenic mouse models
Oncogene | Model | System | Tumor type | Response to inactivation | Mechanism of tumor regression | References |
|---|---|---|---|---|---|---|
BCL2 | MMTV-tTA Tet-O-BCL-2 Eμ-MYC | Tet-off | Lymphoblastic leukemia | Regression | Apoptosis | [142] |
BCR-ABL | MMTV-tTA Tet-O-BCR-ABL | Tet-Off | B-cell leukemia | Regressiona | Apoptosis | [59] |
SCL-tTA Tet-O-BCR-ABL | Tet-Off | CML | Regression | ND | [60] | |
FGF-10 | CCSP-rtTA or SPC-rtTA Tet-O-CMV-FGF10 | Tet-on | Pulmonary adenomas | Regression | ND | [143] |
HER2/NEU | MMTV-rtTA Tet-O-NeuNT | Tet-On | Mammary Carcinomas | Regressiona | Decreased proliferation and apoptosis | [53] |
MET | LAP-tTA Tet-O-MET | Tet-On | Hepatocellular carcinoma | Regression | Decreased proliferation and Apoptosis | [144] |
c-MYC | EμSRα-tTA Tet-O-MYC | Tet-Off | T- and B-cell lymphoma, acute myeloid leukemia | Regressiona | Cell cycle arrest, differentiation and apoptosis | |
EμSRα-tTA Tet-O-MYC | Tet-Off | Osteosarcoma | Regression | Differentiation | [44] | |
MMTV-rtTA Tet-O-MYC | Tet-On | Breast adenoma | Partial regression | ND | ||
LAP-tTA Tet-O-MYC | Tet-Off | Hepatocellular carcinoma | Regression | Apoptosis and differentiation | ||
Plns- MycERTam | Tamoxifen | Pancreatic Islet cell | Regression | Growth arrest, differentiation, cellular adhesion, vascular collapse | [33] | |
Involucrin-MycER Tam | Tamoxifen | Papillomas | Regression | Growth arrest and differentiation | ||
RAS | Tyr-rtTA H-Ras(V12G) Ink4a-/- | Tet-On | Melanoma | Regressiona | Apoptosis, EGFR expression required | |
SP-r-rtTTA RtTA-KiRas(G12C) | Tet-On | Lung adenoma | Regression | ND | [57] | |
CCSP-rtTA Tet-O-KiRas(G12C) | Tet-On | Lung adenoma | Regression | ND | [57] | |
CCSP-rtTA Tet-op-K-Ras4B(G12D) | Tet-On | Lung adenoma | Regression | Apoptosis | [58] | |
Nestin-TVA RCAS-tTA RCAS-Akt RCAS-Tet-O-KRas | RCAS | Glioblastoma | Regression | Apoptosis | [145] | |
WNT | MMTV-rtTA Tet-O-WNT1 P53-/- | Tet | Mammary adenoma | Regressiona | ND | [146] |
The specific consequences of the inactivation of an oncogene depend upon the type of cancer. In some cases, even briefly inactivating an oncogene may be sufficient to induce sustained cancer regression [43, 44], but in other cases, this has not been observed [45]. Oncogene inactivation may uncover the stem cell properties of cancer cells and induce a state of cancer dormancy [45–49]. Finally, the genetic context can affect whether inactivation of an oncogene will induce sustained regression, or if the cancers can relapse acquiring additional genetic events [45, 50–52]. Conditional transgenic models have been generated for many types of signaling molecules implicated in the pathogenesis of neoplasia.
8.3.2.1 Conditional Models of Receptor Induced Tumorigenesis
The Tet system has been used to conditionally overexpress receptors including an oncogenic form of HER2 containing an activating point mutation in its transmembrane domain [52, 53]. Expression was directed to the breast by utilizing the mouse mammary tumor virus (MMTV) promoter to drive the expression of the rtTA protein. Within 4 days of HER2 activation by doxycycline administration, the mice developed hyperplastic abnormalities. Six weeks following oncogene activation, all of the mice developed multiple invasive mammary carcinomas. The tumors were solid invasive carcinomas that often metastasized to the lung. Following 48 h of HER2 inactivation through doxycycline withdrawal, the cancer cells exhibited proliferation arrest and increased apoptosis. The primary carcinomas rapidly and completely regressed in over 90 % of the mice with a mean regression time of 17 days. Within 30 days, the pulmonary metastases had also completely and rapidly regressed. Thus, tumorigenesis appears to be reversible.
However, a majority of the cancers that had a complete regression upon HER2 repression eventually reoccurred. When the primary cancers and metastases were transplanted into syngeneic hosts, they completely regressed only 55–70 % of the time. The relapse tumors all uniformly lacked both endogeneous and transgene protein expression indicating that the cancers had all become HER2 independent [53]. Subsequently, Snail, a transcriptional repressor was found to be activated in relapsed cancers [52]. Therefore, although oncogene inactivation can cause cancer regression, some transgenic tumors are capable of becoming independent of their initiating oncogenic event.
8.3.2.2 GTPases and Cancer Regression
The Tet-On system has been used to generate a conditional model of mutant H-RAS induced melanomas [54, 55]. The tyrosinase gene promoter (Tyr) was used to conditionally overexpress an H-RAS bearing an activating point mutation (V12G) in an Ink4a-deficient background. Approximately 25 % of the mice developed melanomas within 60 days of H-RAS activation. The melanomas were invasive, highly vascular, and notably amelanotic. The tumors exhibited expression of tyrosinase-related-protein-1 (TRP-1), an early melanocyte-specific maker. Within 48 h of H-RAS inactivation through doxycycline withdrawal, the cancers decreased their proliferation and exhibited robust apoptosis. Within 14 days of H-RAS inactivation, the cancers had completely regressed with only microscopically detectable scattered cancer foci. Notably, melanomas transplanted into SCID hosts also regressed upon inactivation of mutant H-RAS. Approximately 30 % of the melanomas resumed growth, even in the absence of H-RAS, but relapsed tumors failed to express TRP-1, suggesting that these tumors were phenotypically different from the primary cancers [55]. Additionally studies have shown that EGFR signaling is required for maintenance of a tumorigenic phenotype in H-RAS induced melanomas. A dominant-negative EGFR reduced the tumorigencity of melanomas, and sustained expression of EGFR can delay cancer regression [56].
The Tet-System has also been used to generate conditional models of mutant K-RAS induced lung adenocarcinoma [57, 58]. The Clara cell secretory protein (CCSP) promoter was used to regulate gene expression in alveolar epithelial cells. Within 7–14 days post induction of K-RAS overexpression, type II pneumocytes exhibited focal hyperplasia, and after 2 months, multiple solid adenomas or adenocarcinomas were present in the lung. The solid adenomas contained a population of macrophages, but lacked invasive growth and stromal elements. The adenocarcinomas had fewer macrophages and cytoplasmic inclusions, but had local invasion of the pleura. Within 3 days of K-RAS inactivation through doxycycline withdrawal, cancers exhibited decreased cellular density and an increased rate of apoptosis. Within 7 days of K-RAS inactivation, only a few patches of hyperplasia were found, and within a month no residual cancer tissue was found in 5/5 mice. The same mice were generated in either a p53 or an Ink4A/Arf deficient background. Cancers grew rapidly in these mice after K-RAS induction, but regressed with the same kinetics. TUNEL assays revealed that regardless of the genetic context cancers were regressing associated with apoptosis [58]. Similarly, CCSP regulated K-RAS (G12C) induced lung adenomas regressed upon oncogene inactivation [57]. Hence, even aggressive lung cancers in a tumor suppressor-deficient background regress upon the inactivation of a single oncogene.
8.3.2.3 Protein Kinases and Cancer Regression
A conditional transgenic model for BCR-ABL leukemias was generated using either the MMTV or SCL (stem cell leukemia) promoter to drive the expression of tTA [59, 60]. Upon induction of BCR-ABL the mice developed B-cell leukemia associated with lymphadenopathy, splenomegaly, and bone marrow infiltration. A third of mice with BCR-ABL under the control of the SCL promoter developed B-cell lymphoblastic disease resembling blast crisis, closely mimicking what is observed in patients with chronic myelogenous leukemia (CML). Inactivation of BCR-ABL induced rapid cancer regression in all mice. BCR-ABL inactivation was associated with the apoptosis of 80 % of the cancer cells within 20 h and complete tumor regression within 5 days. Sustained regression of cancer was observed in tumors arising from three of the four founder lines, as long as the mice had BCR-ABL continuously inactivated. Upon reactivation of BCR-ABL the cancers rapidly recurred. Interestingly, all the mice derived from the fourth founder relapsed within 4 weeks after complete regression. Relapsed cancers lacked continued expression of BCR-ABL protein and mRNA, suggesting that they had become independent of BCR-ABL expression.
8.3.2.4 Nuclear Transcription Factors
The Tet and Tamoxifen systems have been used to demonstrate that MYC inactivation can induce cancer regression in a multitude of different types of cancer (Table 8.1). The Tet-Off System was used to regulate human c-MYC in lymphoid cells under the regulation of the EμSRα promoter [61–63]. When MYC is constitutively activated, 100 % of the mice developed hematopoietic tumors within 5 months. Upon gross examination, the mice exhibited enlargement of the thymus, liver, spleen, and gastrointestinal lymph nodes. Histological examination revealed that cancer cells had invaded all hematopoietic organs as well as liver, kidney, blood, and the lamina propia of the intestines. In one study, tumors were generally immature CD+4/CD+8 T-cell lymphomas and were rarely acute myeloid leukemias [61]. In another study, tumors were either B-cell or T-cell lymphomas [63]. In both studies, the resulting hematopoietic tumors exhibited a high degree of genomic instability reflected by chromosomal gains, losses, or translocations [61, 63]. Despite this genomic complexity, the inactivation of MYC resulted in rapid and sustained cancer regression. Upon MYC inactivation, cancer cells arrested, differentiated, and underwent apoptosis. Over 50 % of tumors exhibit sustained regression for over 30 weeks. Thus, MYC inactivation can induce sustained regression of hematopoietic tumors.
Conditional transgenic mice expressing c-MYC under the control of the Eμ-promoter occasionally developed highly metastatic osteosarcomas [44]. Histological examination of the primary cancer reveals the presence of disorganized bone matrix. MYC inactivation induced rapid cancer regression associated with the differentiation of cancer into mature bone. Continuous video time-lapsed microscopy (CVTL) revealed that upon MYC inactivation, cancer cells ceased to proliferate and differentiated, as confirmed by immunocytochemical analysis. Identically, MYC inactivation in cancers in vivo was associated with the differentiation of malignant cells into mature osteoid. Upon MYC reactivation less than 1 % of the cells were able to regain a proliferative phenotype. Surprisingly, MYC reactivation was also associated with the apoptosis of the now differentiated cancer cells. Moreover, even the transient inactivation of MYC was found to increase the survival of mice with these cancers. Hence, at least in some circumstances even brief oncogene inactivation can induce sustained loss of a neoplastic state.
The tamoxifen system also has been used to evaluate the consequences of MYC inactivation in different types of tumors using MycERTAM (Table 8.2). MycERTAM has been expressed in the skin through the involucrin promoter [34, 43]. MYC activation resulted in increased proliferation and blocked differentiation of the suprabasal epidermis. Sustained MYC activation resulted in hyperplasia, dysplasia, angiogenesis, and papillomatosis. MYC inactivation resulted in regression of blood vessels, restoration of cellular differentiation, and the regression of papillomas. A brief inactivation of MYC in keratinocytes caused the cells to differentiate and become unresponsive to MYC reactivation. MYC reactivation could not restore a proliferative phenotype to the differentiated keratinocytes and eventually the cells were sloughed off the skin [43]. Hence, brief inactivation of MYC can induce the sustained loss of neoplastic features in some skin tumors.
Table 8.2




Oncogene inactivation in the therapeutic setting
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree