Neoplasm
Translocation
Gene/alteration
Alveolar rhabdomyosarcoma
t(2;13)(q35;q14)
PAX3-FOXO1
t(1;13)(p36;q14)
PAX7-FOXO1
t(X;2)(q13;q35)
PAX3-AFX
Alveolar soft part sarcoma
der(17)t(X;17)(p11;q25)
ASPL-TFE3
Angiomatoid fibrous histiocytoma
t(12;22)(q13;q12)
EWSR1-ATF1
t(2;22)(q33;q12)
EWSR1-CREB1
t(12;16)(q13;p11)
FUS-ATF1
Clear cell sarcoma
t(12;22)(q13;q12)
EWSR1-ATF1
t(2;22)(q33;q12)
EWSR1-CREB1
Congenital/infantile fibrosarcoma
t(12;15)(p13;q25)
ETV6-NTRK3
DFSP
t(17;22)(q21;q13) derivative ring chromosomes
COLIA1-PDGFB
Desmoid tumor (fibromatosis)
Trisomy 8 or 20; loss of 5q21
CTNNB1 or APC mutation
Desmoplastic small round cell tumor
t(11;22)(p13;q12)
EWS-WT1
Epithelioid hemangioendothelioma
t(1;3)(p36;q25)
WWTR1-CAMTA1
Epithelioid sarcoma (proximal type)
Inactivation of INI1
INI1
Ewing’s sarcoma/PNET family
t(11;22)(q24;q12)
EWS1-FLI1
t(21;22)(q22;q12)
EWS1-ERG
t(7;22)(p22;q12)
EWSR1-ETV1
t(2;22)(q33;q12)
EWSR1-FEV
t(17 ;22)(q12 ;q12)
EWSR1-E1AF
inv(22)(q12 ;q12)
EWSR1-ZSG
t(16 ;21)(p11 ;q22)
FUS-ERG
Extrarenal rhabdoid tumor
Inactivation of INI1
INI1
Extraskeletal myxoid chondrosarcoma
t(9;22)(q22;q12)
EWSR1-NR4A3
t(9;17)(q22;q11)
TAF2N-NR4A3
t(9;15)(q22;q21)
TCF12-NR4A3
t(3;9)(q11;q22)
TFG-NR4A3
GIST (Sporadic and familial)
Activating kinase mutations
KIT or PDGFRA
ETV1 overexpression
BRAF mutation
Inflammatory myofibroblastic tumor
t(1;2)(q22;p23)
TPM3-ALK
t(2:19)(p23;p13)
TPM4-ALK
t(2;17)(p23;q23)
CLTC-ALK
t(2;2)(p23;q13)
RANBP2-ALK
t(2;11)(p23;p15)
CARS-ALK
inv(2)(p23;q35)
ATIC-ALK
Leiomyosarcoma
MYOCD amplification
Lipoma
t(3;12)(q27-28,q14-15)
HMGA2-LPP
Low-grade fibromyxoid sarcoma
t(7;16)(q33;p11)
FUS-CREB3L2
t(11;16)(p11;p11)
FUS-CREB3L1
Myoepithelioma
t(1;22)(q23;q12)
EWSR1-PBX1
Myoepithelial carcinoma
t(19;22)(q13;q12)
EWSR1-ZNF444
Myxoid/round cell liposarcoma
t(12;16)(q13;p11)
FUS-DDIT3
t(12:22)(q13;q12)
EWSR1-DDIT3
PIK3CA mutation
Myxofibrosarcoma
NF1 deletion, point mutation and indel
Pericytoma
t(7;12)(p22;q13)
ACTB-GLI1
Pleomorphic liposarcoma
NF1 deletion, point mutation and indel
Synovial sarcoma
t(X;18)(p11;q11)
SS18-SSX1
t(X;18)(p11;q11)
SS18-SSX2
t(X;18)(p11;q11)
SS18-SSX4
TGCT/PVNS
t(1;2)(p13;q35)
CSF1-COL6A3
WDLPS/ALT and DDLPS
12q14-15 (supernumerary ring chromosomes ; giant marker chromosomes)
Amplification of MDM2 , CDK4, HMGA2, SAS, GLI and JUN
Cytogenetic analysis of alveolar rhabdomyosarcoma, a common soft tissue neoplasm of young children, has demonstrated two specific chromosomal translocations [8]: the t(2;13)(q35;q14) translocation, which is found in 60 % of cases, and a less common variant t(1;13)(p36;q14) [9, 10]. These involve the FKHR gene (FOXO1A) on chromosome 13q14 and either Pax3 (2q35) or Pax7 (1p36) genes [11]. The Pax7-FKHR tumors are often seen in younger patients and are associated with a lower rate of metastasis and better survival than Pax3-FKHR tumors [12]. There is no specific chromosomal abnormality identified involving the embryonal subtype of rhabdomyosarcoma [13].
Desmoplastic small round cell tumor demonstrates multilineage differentiation and characteristically possesses the t(11;22)(p13;q12) translocation that fuses the EWSR1 gene to WT1, the Wilm’s tumor suppressor gene, on chromosome 11p13 [4, 14].
Extraskeletal myxoid chondrosarcoma, an uncommon sarcoma of uncertain lineage, demonstrates the EWS-NR4A3 fusion generated from a t(9;22)(q22;q12) translocation [15]. A second translocation that may be encountered is t(9;17)(q22;q11) [16]. Of note, there is no immunohistochemistry profile that can reliably diagnosis this particular sarcoma, making molecular studies important for the diagnosis of extraskeletal myxoid chondrosarcoma [17].
In addition to the small round blue cell tumors, several spindle cell sarcomas are associated with recurrent translocations. Importantly, synovial sarcoma, an aggressive neoplasm of uncertain histogenesis, demonstrates a recurrent reciprocal t(X;18)(p11.2; q11.2) translocation [1, 18]. The translocation fuses the SYT gene on chromosome 18q11 to one of the three homologous genes on Xp11 (SSX1, SSX2, or SSX4), which is detectable in 95 % of synovial sarcomas [2, 19]. The SYT-SSX1 fusion transcript is reportedly associated with biphasic histology, whereas the SYT-SSX2 fusion transcript is associated with a monophasic histology as well as a better metastasis-free survival [20].
Dermatofibrosarcoma protuberans (DFSP), a low-grade spindle cell neoplasm, possesses a specific chromosomal rearrangement consisting of either supernumerary ring chromosomes or unbalanced derivatives of t(17;22)(q22;q13) translocation. Both aberrations have sequences encoding a COL1A1-PDGFB fusion transcript that is commonly identified by reverse transcription-polymerase chain reaction (RT-PCR) [21–23]. Reportedly, imatinib mesylate (Gleevec, STI571/Glivec) has clinical activity against DFSP with t(17;22); however, fibrosarcomatous variants of DFSP without the translocation may not show a response. Molecular testing would be important in identifying patients with DFSP who have the translocation because they would more likely respond to therapy [24].
Congenital fibrosarcoma, another spindle cell sarcoma, has a characteristic cytogenetic change, t(12;15)(p13;q25) translocation, not found in the adult fibrosarcomas. In addition, the chromosomal rearrangement is identical to that described for congenital mesoblastic nephroma, acute myeloblastic leukemia, and secretory carcinoma of the breast [25–27]. Fusion of the ETV6 gene on 12p13 to the neurotrophin 3 receptor gene (NTRK3 or TRKC) on chromosome 15q25 is often difficult to identify cytogenetically and is better detected by FISH or PCR [28, 29].
Low-grade fibromyxoid sarcoma, a rare variant of fibrosarcoma, is associated with the FUS-CREB3L2 gene fusion or less frequently the FUS-CREB3L1 gene fusion, the result of translocations t(7;16) or t(11;16), respectively [30, 31]. The chromosomal abnormalities may be detected with either FISH or RT-PCR [32].
Gastrointestinal stromal tumor (GIST), a spindle cell sarcoma, is unlike the previous spindle cell sarcomas discussed in that it is characterized by activating oncogenic mutations instead of specific translocations or fusion genes [3]. KIT mutations are a defining feature of most GISTs, and, to a lesser degree, mutations in platelet-derived growth factor receptor alpha (PDGFRA) can also occur in GIST. These activating mutations lead to constitutive activation of their respective kinases, leading to downstream effects that decrease apoptosis and promote cellular survival [33–36]. A majority of GISTs reveal a mutation in one of three sites: KIT exon 11, KIT exon 9, or PDGFRA exon 18 [35, 37]. These mutation types correlate with the anatomic location and have prognostic implications. The KIT exon 11 mutations include in-frame deletions, point mutations, and internal tandem duplications; however, patients with point mutations fare better prognostically than those with deletions [38]. Small intestinal GISTs usually demonstrate KIT 9 exon mutations, and patients appear to have a higher mortality with small intestinal versus gastric GISTs with KIT 9 exon mutations [36, 39]. PDGFRA mutations are primarily localized to the stomach and have an epithelioid morphology, and they frequently exhibit an indolent clinical course [40, 41]. GISTs have been successfully treated with tyrosine kinase inhibitors that specifically target c-Kit, such as imatinib mesylate and sunitinib, which further support molecular genotyping of these tumors because the genotype correlates with different treatment outcomes, including progression-free survival and overall survival [35, 42–44]. The majority of laboratories use PCR to amplify the most commonly mutated exons with subsequent direct sequencing analysis of the amplified exon [3].
There are a few notable chromosomal abnormalities and genetic aberrations involving lipomatous neoplasms. Myxoid-round cell liposarcoma is known for its characteristic t(12;16)(q13;p11) reciprocal translocation, resulting in the formation of FUS-DDIT3 chimeric gene in 95 % of cases [4, 45, 46]. Myxoid-round cell liposarcoma is radiosensitive; with the use of molecular studies as an adjunct to histopathology, patients will be able to get neoadjuvant therapy and reduce the risk of an ablative procedure [32]. Recently, Barretina et al. [47]performed a high-throughput, integrative analysis of DNA, mRNA, and mutational data to show that 18 % of myxoid-round cell liposarcomas contain mutations in the phosphotidylinosital-3 kinase catalytic alpha polypeptide (PIK3CA). Compared to the wild-type version, tumors with PIK3CA exhibited a shorter progression-free survival, suggesting a more aggressive course. On the other hand, well-differentiated liposarcoma/atypical lipomatous tumor (WDLPS/ALT) cytogenetically shows supernumerary ring chromosomes, giant marker chromosomes, and double minutes in 80 % of cases, corresponding to the amplification of the 12q13-15 band [2]. Several tumor-associated genes are localized to that band region, including MDM2 and CDK4 [48]. Pleomorphic liposarcomas, a less common liposarcoma variant, does not exhibit a specific chromosomal translocation, but these tumors have been shown to exhibit mutations in TP53 and NF1 [47].
There are sarcomas of uncertain histogenesis with characteristic translocations. Clear cell sarcoma (malignant melanoma of soft parts) demonstrates t(12;22)(q13;q12) translocation involving the ATF-1 gene on chromosome 12q13 and EWSR1 [49, 50]. Alveolar soft part sarcoma (ASPS) has a recurrent der(17)t(X;17)(p11;q25) unbalanced translocation [51]. This translocation involves the fusion of TFE3 transcription factor gene on Xp11 and the ASPL gene on chromosome 17q25 [52]. The ASPSCR1-TFE fusion protein has been associated with overexpression of MET, and inhibition of MET by RNA interference has resulted in decrease growth of ASPS cells lines [53].
Extrarenal (and renal) rhabdoid tumors, proximal-type epithelioid sarcoma, and a subset of pediatric undifferentiated soft tissue sarcomas have an inactivating oncogenic mutation, specifically an inactivation of the tumor suppressor gene SMARCB (INI1) [54–57]. Basically, there is either a loss of both alleles or loss of one allele with an inactivating mutation of the other, resulting in the inactivation of the tumor suppressor gene [3]. SMARCB molecular abnormalities may be detected using conventional karyotyping, FISH, and direct sequencing and more recently with multiplex ligation-dependent probe amplification of single nucleotide polymorphism-array analysis to identify small deletions, duplications, or single base pair mutations within the tumors [3, 58].
13.3 Bone Tumor-Specific Genetic and Molecular Abnormalities
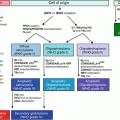
Approximately 0.2 % of bone tumors are primary, which is less frequent than metastasis to the bone and less common than soft tissue counterparts (1:10). There are 19 main subtypes of benign bone tumors and 22 subtypes of bone sarcomas. Table 13.2 summarizes the most common and recent cytogenetic and molecular abnormality of bone neoplasms.
Table 13.2
Characteristic genetic alterations in bone neoplasms
Neoplasm | Translocation | Gene/alteration |
|---|---|---|
Aneurysmal bone cyst | t(16;17)(q22;p13) | CDH11-USP6 |
t(1;17)(p34.3;p13) | THRAP3-USP6 | |
t(3;17)(q21;p13) | CNBP-USP6 | |
t(9;17)(q22;p13) | OMD-USP6 | |
t(17 ;17)(q21 ;p13) | COL1A1-USP6 | |
Bizarre parosteal osteochondromatous proliferation (Nora lesion) | t(1;17)(q32 ;q21) | RDC1 |
Chondrosarcoma or chondroma | IDH1 or IDH2 | |
Point mutation | ||
Ewing’s sarcoma/PNET family | t(11;22)(q24;q12) | EWS1-FLI1 |
t(21;22)(q22;q12) | EWS1-ERG | |
t(7;22)(p22;q12) | EWSR1-ETV1 | |
t(2;22)(q33;q12) | EWSR1-FEV | |
t(17;22)(q12;q12) | EWSR1-E1AF | |
inv(22)(q12;q12) | EWSR1-ZSG | |
t(16;21)(p11;q22) | FUS-ERG | |
Fibrous dysplasia | Activating oncogenic mutations | GNAS1 |
Mesenchymal chondrosarcoma | t(8;8)(q13;q21) | HEY1-NCOA2 |
Osteosarcoma, low grade (parosteal and intramedullary) | 12q14-15 (ring chromosomes, giant marker chromosomes) | Amplification of CDK4, MDM2 , HMGA2, GLI and SAS |
Osteosarcoma or chordoma | Multiple structural rearrangements | |
Subungual exostosis | t(X;6)(q24-q26;q15-21) | COL12A1-COL4A5 |
Bone tumors are primarily of mesenchymal origin. Tumors of the bone as classified by the World Health Organization may be cartilaginous, osteogenic, fibrogenic, fibrohistiocytic, vascular, myogenic, lipogenic, neural, or notochordal [1]. However, there are instances when bone is involved by tumors of neuroectodermal origin, such as Ewing Sarcoma, or by tumors of undefined histogenesis, such as aneurysmal bone cyst.
Cartilagenous tumors produce a chondroid matrix and may either be benign or malignant in nature. Osteochondroma, a cartilage capped bony projection arising on the external surface of bone containing a marrow cavity continuous with that of the underlying bone, may form as a solitary lesion or multiple lesions as in hereditary multiple osteochondromas (HMO) [1]. Cytogenetic aberrations have been identified in the EXT1 gene located at 8p22-24.1 in sporadic and hereditary osteochondromas. Specifically, germline mutations in the tumor suppressor genes EXT1 located at 8q24 and EXT2 located at 11p11-p12 have been found in HMO [59–61]. Enchondroma is a benign hyaline cartilage tumor of medullary bone commonly involving the hands and feet [1]. By conventional cytogenetic analysis, aberrations involving chromosome 12 have been detected [62, 63]. Chondromyxoid fibroma frequently occurs in the long bones, particularly the tibia. It is a benign tumor composed of lobules of spindle-shaped cells with abundant myxoid or chondroid intercellular material [1]. Structural rearrangements of chromosome 6 have been found to be non-random, specifically involving the long arm (q13 and q25) and short arm (p25) [64–68]. Chondrosarcoma is a malignant tumor of hyaline cartilage differentiation that may contain myxoid changes, calcifications, or areas of ossification [1]. There are several subtypes of chondrosarcoma, including conventional, periosteal, dedifferentiated, mesenchymal, and clear cell. Genetic alterations involving chondrosarcoma are heterogeneous. Comparative genomic hybridization studies have revealed extensive genetic alterations, including gains of whole chromosomes or chromosome arms at 20q, 20p, and 14q23 or partial losses [69]. Loss of genetic material from 13q was found to be an independent predictor of metastasis development, regardless of tumor grade or size [68, 70, 71]. Dedifferentiated chondrosarcoma, a subtype of chondrosarcoma composed of a well-differentiated cartilage tumor juxtaposed to a high-grade non-cartilaginous sarcoma; reportedly possesses frequent structural and numerical alterations of chromosome 1 and 9 [1, 66, 72–75]. Identical somatic deletion in exon 7 of p53 have been identified in the cartilaginous, as well as within the dedifferentiated part [1, 66]. Mesenchymal chondrosarcoma is a rare subtype characterized by a biphasic pattern composed of undifferentiated small round cells and islands of well-differentiated hyaline cartilage [1]. In two cases of mesenchymal chondrosarcoma a Robertsonian (13;21) translocation was observed [76].
Osteogenic tumors produce a bony matrix or what is referred to as ‘osteoid’ and, depending on their behavior, are considered either benign or malignant. However, rarely benign osteogenic tumors undergo malignant transformation [1]. Osteoid osteoma is a benign cortically based bone-forming tumor defined by its small size, limited growth potential (seldom exceeds 1.0 cm), and disproportionate pain. Reportedly, there have been two of three cases of osteoid osteoma in which a loss of the distal part of chromosome arm 17q was detected [77]. Osteosarcoma, a highly malignant bone tumor, commonly arises within the long bones of children; however, about 15 % arise in adults secondary to a pre-existing condition, such as Paget disease [1]. The classical or conventional osteosarcoma may histologically show a variable of features and be subtyped as either osteoblastic, chondroblastic, or fibroblastic. There are other more rare subtypes, including telangiectatic osteosarcoma, small cell osteosarcoma, parosteal osteosarcoma, periosteal osteosarcoma, high-grade surface osteosarcoma, and more. Most osteosarcomas possess complex chromosomal alterations. However, there appear to be certain chromosomal regions that display recurrent amplifications. Amplification at 1q21-23 and 17p, together with co-amplification of 12q13-15, are frequently reported [78–84]. The MDM2 gene is amplified in 14–27 % of osteosarcomas [82, 85, 86]. Amplification of CDK4 gene is found in aggressive lesions [87–89]. Lastly, the MYC gene has been shown to be amplified in 44 % of osteosarcomas (8q24) [90]. Understandably, low-grade variants of osteosarcoma have not been found to harbor complex aberrations. For example, low-grade central osteosarcoma show recurrent gains in minimal common regions at 12q13-14, 12p, and 6p21 [91]. Parosteal osteosarcoma, another low-grade osteosarcoma that arises on the surface of bone, reportedly shows ring chromosomes with co-amplification of SAS, CDK4, and MDM2 genes with a minimal common region at 12q13-15 [92–94]. As stated previously, osteosarcomas may arise secondary to a pre-existing condition (Paget disease) or after radiation therapy. In tumors forming after radiation therapy, chromosomal losses are more often observed than gains [95]; 1p is lost in 57 % of post-radiation osteosarcomas, in contrast to conventional osteosarcomas that demonstrate only a 3 % loss [95, 96]. Osteosarcomas arising secondary to Paget disease of the bone show a frequent loss of heterozygosity at 18q [97]. A unique and rare tumorous lesion with aggressive growth known as bizarre parosteal osteochondromatous proliferation (Nora’s lesion) is an exception to the rule, in that this neoplasm possesses a unique recurrent aberration t(1;17)(q32;q21) not found in most bone-forming tumors [98].
Fibrogenic tumors do not produce a mineralized matrix, but form collagen. Desmoplastic fibroma of the bone is a rare, benign neoplasm composed of spindle cells with minimal atypia and abundant collagen [1]. The histomorphology is identical to fibromatosis of the soft tissue. Like its soft tissue counterpart, a subset of desmoplastic fibromas of the bone show trisomies of 8 and 20 [99].
Ewing’s sarcoma, as previously mentioned above, demonstrates the same aberrations as seen in its soft tissue counterpart.
Giant cell tumor of the bone is a locally aggressive neoplasm. Histology reveals sheets of neoplastic ovoid mononuclear cells interspersed with uniformly distributed large osteoclast-like giant cells [1]. A frequent chromosomal aberration detected in giant cell tumor of the bone is a reduction in the length of telomeres. Most commonly affected telomeres are 11p, 13p, 14p, 15p, 19q, 20q, and 21p [100–105].
Chordoma is a neoplasm that arises from notochordal remnants. It usually occurs in the sacrum or clivus. Typically, chordomas display a lobular growth pattern and contain neoplastic cells with bubbly cytoplasm known as “physaliphorous cells” [1]. In 9 of 16 cases of chordomas, hypodiploidy was identified. Chromosomes 3, 4, 10, and 13 were frequently lost [68, 106]. A tumor suppressor gene is believed to exist on distal 1p due to loss of heterozygosity at band 1p36 [107].
Admantinoma, a malignant biphasic tumor characterized by an epithelial and osteofibrous component, may derive from their osteofibrous dysplasia-like counterparts. Osteofibrous dysplasia and adamantinoma show similar cytogenetic alterations such as trisomies of chromosomes 7, 8, 12, 19, and 21 [108–111].
As described in soft tissue tumors, there are also tumors of undefined histogenesis that occur in the bone. Primary aneurysmal bone cyst, a cystic lesion of bone composed of blood-filled spaces separated by connective tissue septa with fibroblasts, giant cells, and reactive woven bone, has been shown to harbor a recurrent translocation t(16:17) in several cases [1, 112]. However, rearrangements are absent in secondary aneurysmal bone cysts [113]. Over the years, additional translocations have been identified, all of which result in oncogenic activation of the USP6 gene on chromosome 17p13 [114–116]. Fibrous dysplasia, a benign medullary fibro-osseous lesion, may involve one (monostotic) or more bones (polyostotic) [1]. It is characterized by activating mutations in the GNAS1 gene located on chromosome 20q12–q13.3, encoding the alpha subunit of the stimulatory G protein [117].
13.4 Therapy for Soft Tissue Tumors
Benign tumors are generally treated with surgical excision. Malignant tumors (sarcomas) can be treated with combination therapy, including surgery, chemotherapy, targeted therapy, radiation therapy, hormonal therapy, and immunotherapy. Surgery achieves local control and removes tumor burden. Radiation therapy is applicable when the surgical margin is positive or used neoadjuvant to facilitate subsequent resection. Cytotoxic chemotherapy is used for metastatic disease, unresectable tumors, or sensitive histological types. Sensitivity to chemotherapy is variable among sarcomas, and, at this time, histology appears to be the best predictor of response. Chemotherapy-sensitive tumors include Ewing’s sarcoma, rhabdomyosarcoma, myxoid/round cell liposarcoma, synovial sarcoma, uterine leiomyosarcoma, desmoid tumor (fibromatosis), desmoplastic small-round-cell tumor, and angiosarcoma. Resistant histologic types include ASPS, clear cell sarcoma, and extraskeletal myxoid chondrosarcoma. Dedifferentiated liposarcomas, epithelioid sarcomas, and perivascular epithelioid cell tumors are minimally sensitive to cytotoxic chemotherapy, whereas fibrosarcoma, high-grade undifferentiated sarcoma, malignant peripheral nerve sheath tumor, solitary fibrous tumor/hemangiopericytoma, and hemangioendothelioma exhibit intermediate sensitivity to chemotherapy. Although histology may be the best predictor of response, histologic grade is the greatest predictor of recurrence [118]. Unfortunately, there are many more high-grade sarcoma subtypes resistant to anthracycline-based therapies compared to chemotherapy-sensitive subtypes. Furthermore, most clinical trials from the 1970s to present often “lump” multiple histologic subtypes together to increase the total sample size at the expense of diluting the overall effect of the chemotherapeutic agent. Thus, endpoints such as response rate or survival are often modest in benefit, resulting in controversy among many sarcoma specialists. Clearly, there is a need to improve our understanding of the molecular pathology so that the survival of patients afflicted with these rare diseases can be improved.
An excellent example of molecular-based therapy involves GISTs. This rare mesenchymal tumor is postulated to arise from the interstitial cell of Cajal and commonly involves the stomach and small intestine of adults (peak age of 60), with less than 10 % of cases in individuals under age 40 years. They may occur in extra-gastrointestinal sites, including the retroperitoneum, mesentery, and omentum. Ninety percent of GISTs harbor a mutation within the KIT gene and/or PDGFRA gene (80 % and 10 %, respectively) [119]. Recently, V600E BRAF mutation was identified in adult patients with GIST lacking KIT and PDGFRA mutations [120, 121]. GIST mutational analysis is important in the evaluation of patients with GIST tumors to analyze prognosis, to make decisions regarding adjuvant treatment, to predict treatment response, and to select appropriate dosage. Different types of mutations involving different exons result in varying clinical prognosis, histologic appearance, and treatment decisions for patients. Internal tandem repeats involving exon 11 are associated with gastric GISTs and an indolent course, whereas deletions within exon 11 are associated with a shorter survival, higher risk of recurrence, and an aggressive course. Exon 9 mutations are associated with small intestinal GISTs and a clinically aggressive course. PDGFRA missense mutations in exon 18, exon 14, or exon 12 occur in GISTs involving the stomach, which have an epithelioid or mixed epithelioid/spindle cell histology [3, 122, 123]. Historically, treatment of advanced GIST is revolved around surgery, with few effective systemic therapeutic options; however, the introduction of imatinib mesylate (ST1571; Gleevec), a selective tryosine kinase inhibitor that targets KIT and PDGFRA, has changed the course of this disease.
Imatinib is an oral FDA-approved drug for use as an adjuvant therapy following complete gross resection of tumor and in the management of unresectable and metastatic disease. GIST tumors with exon 11 mutations are the most responsive to imatinib therapy and GISTs with exon 9 mutations may benefit from an increase dose (800 mg). Patients with wild-type GIST and wild-type PDGFRA rarely show a favorable or sustained response to treatment. In this situation, other less common gene mutations are likely involved, such as in the succinyl dehydrogenase gene, noted in patients with an inherited form of GIST that is often accompanied by the development of paragangliomas, and pulmonary hamartomas [124]. Other forms of wild-type KIT and wild-type PDGFRA GIST include tumors with activation the Ras pathway in NF1 patients or in tumors with insulin-like growth factor 1 receptor (IGF1R) amplification. Unfortunately, there are cases where patients with exon 9 and PDGFRA mutations have primary resistance to imatinib likely due to the structure of the ATP-binding loop, which results in decreased affinity for the drug [125]. Secondary resistance occurs and generally is due to additional mutations involving exon 11. Sunitinib malate (SU11248; Sutent) is a multi-targeted tyrosine kinase inhibitor that also inhibits vascular endothelial growth factor receptors (VEGFRs). It is used to treat patients with primary or secondary resistance to imatinib. Patients with exon 9 mutations or wild-type genotyping demonstrate a good response, although the overall response rate is noted to be less than 10 % [126]. As with imatinib, sunitinib resistance may evolve and thus represents an area of active investigation. Novel therapeutic strategies that target different aspects of the intracellular signaling pathway involving KIT and PDGFRA are being investigated, and potential options include the following: heat-shock protein (HSP)-90 inhibitor, everolimus, nilotinib, sorafenib, dasatinib, regorafenib, masitinib, PI3-kinase/mTOR dual inhibitors, and PI3-kinase inhibitors [127]. In a phase II efficacy study of sorafenib, partial response (13 %) and stable disease (58 %) were reported in 24 patients with resistance to imatinib or sunitinib. A multicenter phase II study of Masitinib revealed preliminary observations of objective partial response (50 %), stable disease (47 %), and primary refractory (3 %) with overall disease rate of 97 %. Phase III studies are underway comparing nilotinib and masitinib as single agents with imatinib in the first-line setting. The HSP-90 inhibitor inhibits HSP-90, a chaperone protein that aids in protein folding to stabilize KIT from degradation, to prevent stabilization of KIT, and to lead to its degradation [127].
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree