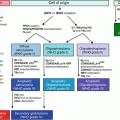
Fig. 3.1
Murine mammary gland development. (a) During embryonic development, bilateral milk lines are established by embryonic day 10 (E10). The milk line breaks up and coalesces into individual mammary placodes which transform into elevated buds by E11. Each bud continually develops and invaginates below the epithelial surface to form a bulb structure that condenses the underlying stroma and pushes into the mammary mesenchyme towards the fat pad. Sprouts branch from the bulb and penetrate into the mammary mesenchyme in formation of a primitive tree like structure by E18. (b) Formation of the mature mammary gland begins in the presence of circulating steroid and peptide hormones systemically produced at the onset of puberty. ( c) Each pregnancy cycle involves the formation of milk producing alveolar sacs. The mammary gland undergoes post-lactational involution after weaning
3.3 ER, PR, HER2/neu, BRCA1/2, Grade, Stage, and Lymph Node Status
Current diagnosis emphasize standard clinical features such as the tumor size, extent of lymph node involvement, and distant metastasis and have all been integrated into the American Joint Committee on Cancer TNM classification standard and still play a central role in determining prognosis and treatment course by the physician [15–20]. Similarly, ER and PR status have been defined as important markers of breast cancer pathology decades ago, but now are gold standards in prescribing adjuvant therapy [21–28]. Another important marker for therapy response of breast cancer is the amplification of HER2/neu. Testing for this marker has become routine before prescribing Herceptin therapy. HER2/neu is a cell membrane surface-bound receptor tyrosine kinase belonging to the epidermal growth factor receptor family (ErbB) involved in signal transduction pathways promoting cell growth and differentiation [29–35]. Although its presence indicates aggressive tumor, trastuzumab, a specific antibody that targets the cell surface receptor, is supplemented, so that the receptor can be blocked from receiving growth signals [36–39].
A subtype of aggressive breast cancer, called “triple negative,” lacks ER/PR/HER2 expression and grows without the need for external growth hormones [40–46]. It is more prevalent in younger women and characterized by early recurrence and visceral metastasis to the brain [40, 47, 48]. Fortunately, it is still sensitive to conventional chemotherapy and often prescribed concurrently with surgery [40, 41, 49–51]. Although the molecular biology of this breast cancer subtype is not yet fully understood, it has been recently associated with mutations in the two major breast cancer susceptibility genes, BRCA1 and BRCA2 [52–56].
BRCA1 and BRCA2 function as tumor suppressors involved in DNA damage response and DNA repair. Mutation in one copy of either BRCA1 or BRCA2 can increase the risk of hereditary breast and ovarian cancer with a lifetime risk of 50–80 % and 30–50 % for breast and ovarian cancer, respectively. Additionally, cancer development occurs earlier than non-hereditary breast cancer and is often aggressive [57].
Other points in early classification schemes are stage, grade, and lymph node status. Tumor size is used to assign stage, whereas the Nottingham histological grading system defines proliferative activity (mitotic index) and degree of differentiation (tubule formation and nuclear pleomorphism) of the tumor [58, 59]. Assessment of lymph node status is also part of tumor staging, and it can provide prognostic value. However, the combination of all three is often used to predict tumor prognosis. It has been reported that late stage and high grade can override the positive effects of negative node involvement [15, 17, 60–64].
Although determining ER, PR, and HER2 status may seem trivial and often conflicts with classification schemes, it is significant since it allows for targeted therapy. Targeting the ER and widespread use of the selective ER modulator tamoxifen in breast cancer are well-established targeted therapeutic modalities. These are responsible for the major improvements in cure rates, quality of life, and disease prevention for breast cancer observed over the past two decades. Targeting both HER2/neu with trastuzumab and the vascular endothelial growth factor with bevacizumab in combination with chemotherapy has become a further milestone in molecularly targeted therapy of this disease [65–69].
3.4 Gene Expression Arrays
The Stanford group pioneered class discovery through the use of high-throughput technologies and re-emphasized that breast cancer is not a single entity and that it can be classified into at least five groups by their molecular signature: luminal subdivided into luminal A and luminal B; basal-like; HER2; and a least characterized normal breast-like. Luminal tumors, like normal luminal epithelial cells of the breast, express cytokeratins 8/18, and are ER positive. This group is further subdivided into luminal A (lower grade and excellent prognosis) and luminal B (higher grade and poorer prognosis exemplified by higher proliferation rates). The basal-like breast cancers express genes resembling normal basal/myoepithelial mammary cells. For instance, they express cytokeratins 5, 6, 14, and 17, P-cadherin, caveolins 1 and 2, and EGFR. They are often ER, PR, and HER2/neu negative and are of higher histological grade characterized by a high mitotic index. They exhibit necrotic regions with lymphocytic infiltrate and often metastasize. HER2 tumors are ER negative with high HER2/neu expression. This type of breast cancer is aggressive but, unlike the basal like tumors, responds to adjuvant therapy. The last group is the normal breast-like and is not well characterized, but their gene expression profile resembles fibroadenomas and normal breast samples [4, 70–78].
Although classification based on gene expression arrays may seem more objective and straightforward, there are always exceptions. For instance, not all breast tumors that are HER2 amplified can be categorized into the HER2 group. Some HER2-amplified tumors express ER, and their gene expression signature resembles the luminal type. Furthermore, the basal type is not always triple negative (ER, PR, and HER2), and a small subset of triple negative breast cancers is not basal like. Therefore, the use of microarrays to classify breast cancer might not always align with previous histopathological classifications. Nevertheless, MammaPrint®, Oncotype DX®, and Theros® have been developed in recent years for molecular diagnosis of breast cancer gene expression array technologies, and we provide a brief overview below of each [4, 70–82].
3.4.1 MammaPrint®
The first breast cancer classification through utility of gene expression profiles was demonstrated by van’t Veer et al. [83]. The original data sets were samples from patients younger than 55 years old, tumor size smaller than 5 cm, and lymph node negative. They analyzed the gene expression signature of samples taken from breast cancer patients who developed metastasis within 5 years after diagnosis and compared these against the gene expression signatures of samples taken from breast cancer patients who were metastasis free during the same time. The analysis identified a 70-gene expression signature that could provide a predictive value for 5 years metastasis. This resulted in administration of proper treatment to patients with aggressive breast cancer.
The results from this study were validated in a subsequent study by Vijver et al. that encompassed more patients. They determined this gene set was an independent predictor of outcome in a multivariate survival analysis. Beyond its ability to demonstrate likelihood of metastasis, this gene expression signature could stratify node-positive patients based on disease outcome prediction. This assay is currently undergoing phase III clinical trial organized by the European Organization for Research and Treatment of Cancer. The trial is setup to profile the 70-gene expression signature and to simultaneously determine the clinicopathological characteristics for 6,000 patients. If the 70-gene expression signature and clinicopathological data agree, patients will be treated accordingly. However, if the two diagnostic tools differ, patients will be randomized and treated according to the results of the 70-gene expression signature or clinicopathological parameters [84–89].
Finally, this 70-gene expression signature has been shown to outperform traditional clinical and histological parameters for providing a prognostic value [90–93]. A downside to this gene expression signature is that it fails to stratify ER-negative or HER2-positive breast cancer since it predicts 96–100 % and 78–95 %, of them, respectively, which will have poor prognosis. For the remaining 5–22 % HER2-positive breast cancers not predicted to be in the high-risk group, the controversial question is whether to provide trastuzumab treatment or not [93–95]. Regardless, this assay received FDA approval for use in patients younger than 61 years old, at stages I/II, node negative, and tumor size smaller than 5 cm [84–89].
3.4.2 Veridex®
In the original study, Wang et al. [96] separated 115 lymph node-negative breast cancer samples, from systemic therapy naïve patients, into 35 ER-negative and 80 ER-positive for analyses. From the analyses, samples from patients who were ER-positive with distant metastasis expressed 16 genes differentially, whereas samples from patients who were ER-negative with distant metastasis expressed 60 genes differentially. This resulted in a 76-gene signature that can predict likelihood of metastasis. A subsequent study expanded to examine 171 lymph node-negative tumors from systemic therapy-naïve patients. This gene expression signature was shown to be an independent prognostic factor in multivariate analysis. It should be noted that this gene expression signature failed to provide a predictive value when applied to ER-negative patients [96–99].
3.4.3 Theros®
Ma et al. analyzed the gene expression signatures of 60 patients uniformly treated with adjuvant tamoxifen alone. They analyzed the whole tissue sections and laser-capture microdissected samples from patients who responded to tamoxifen treatment and identified three strongly predictive genes: homeobox gene HOXB13, interleukin 17B receptor (IL17BR) and EST AI240933. The study showed expression ratios between HOXB13 and IL17BR were strongly correlated with recurrence and outperformed other clinical pathological predictors. A validation study was carried out from RNA samples extracted from paraffin-embedded tissues from a cohort of 20 patients. Then the HOXB13:IL17BR expression ratio was re-evaluated in a cohort of 206 postmenopausal patients with ER-positive breast cancer who underwent tamoxifen-only treatment from the North Central Cancer Treatment Group (NCCTG) 89-30-52 trial. It was further expanded into a study with a cohort of 852 patients. In these studies, the HOXB13-to-IL17BR ratios were shown to identify ER-positive breast cancer patients that underwent tamoxifen treatment but at high risk of recurrence. It was also shown that the HOXB13-to-IL17BR ratios were a predictor of outcome for ER-positive patients who were either adjuvant naïve or who underwent tamoxifen treatment. Although the HOXB13:IL17BR ratios are practical in these aspects, it appears to have limited ability to predict outcome in lymph node-positive patients. These studies have led to the development of Theros®, a RT-qPCR-based test performed in a central laboratory and commercialized by Biotheranostics. This test is recommended for lymph node negative ER-positive breast cancers patients treated with surgery alone, to define risk of recurrence and benefit from endocrine therapy. At the time of this review, the HOXB13:IL17BR index has not yet been shown to predict adjuvant hormone therapy or chemotherapy benefit and has not been included in the American Society of Clinical Oncology or National Comprehensive Cancer Network guidelines for breast cancer treatment [100–104].
3.4.4 Oncotype DX®
The OncotypeDX® platform (Genomic Health Inc., Redwood City, CA, USA) utilizes RT-qPCR to analyze the expression of 16 cancer-related genes: Ki67, STK15, survivin, CCNB1, MYBL2, GRB7, HER2, ER, PGR, BCL2, SCUBE2, MMP11, CTSL2, GSTM1, CD68, and BAG1, in addition to five reference genes (ACTB, GAPDH, RPLPO, GUS, and TFRC). The test utilizes RNA extracted from formalin-fixed paraffin-embedded ER-positive breast cancer samples. Selection of genes was originally derived from published literature, genomic databases, and microarray data.
OncotypeDX® predicts risk of recurrence in ER-positive, lymph node-negative breast cancer patients and provides treatment guidelines via a quantitative system of continuous Recurrence Score (RS) that ranges between 0 and 100 to predict the risk of recurrence within 10 years. This score has also been shown to be an independent predictor of outcome in multivariate survival analyses where samples are categorized, based on the RS into three risk group: low (RS < 18), intermediate (RS = 18–31), and high (RS ≥ 31).
Studies have validated OncotypeDX® and demonstrated that breast cancer patients who are ER-positive with a low RS tend to have a low risk of recurrence and will not benefit much from chemotherapy. On the other hand, breast cancer patients who are ER-positive with a high RS tend to have a high risk of recurrence and can benefit from chemotherapy. This justifies the use of endocrine therapy in addition to chemotherapy for ER-positive patients with a high RS score. This assay has been implemented into the American Society of Clinical Oncology guidelines for the use of tumor markers in breast cancer and into the National Comprehensive Cancer Network guidelines for breast cancer treatment [105–110].
3.5 MicroRNAs
The miRNA paradigm shifted from worm development to cancer when G.A. Calin et al. identified miR-15 and miR-16 as tumor suppressors at 13q14 in chronic lymphocytic leukemia in 2002 [111]. It has been shown that hemizygous or homozygous loss of 13q14 occurs in more than 50 % of B-cell chronic lymphocytic leukemia, mantle cell lymphoma, and prostate cancers and only slightly lower in multiple myeloma; however, detailed genetic analysis of traditional genes located in this region yielded inconclusive evidence. The discovery of miRNA prompted Calin et al. to revisit information coded within this region. They discovered that the genetic code for two miRNAs, miR-15 and miR-16, are indeed located in this area and hypothesized that deletion of 13q14 results in loss of miR-15 and miR-16 expression. This was confirmed when analyses showed 68 % of RNA samples from B-cell chronic lymphocytic leukemia patients do not express these two miRNAs [111]. Shortly after, Michael et al. showed a similarly reduced expression of mature miR-143 and miR-145 in colorectal cancer [112]; Metzler et al. showed an accumulation of miR-155 in Burkitt lymphoma [113], and a similar trend was reported for other types of B-cell lymphomas, including diffuse large B-cell lymphoma [114, 115]. In 2004, Takamizawa et al. showed that let-7 was often lost in lung cancer and had a direct correlation to postoperative survival. They also found that overexpression of exogenous let-7 in lung cancer cell lines leads to growth inhibition [116]. Later, to complement Takamizawa’s findings, Johnson et al. published data demonstrating that let-7 is indeed a probable tumor suppressor by targeting oncogenic RAS [117]. These landmark studies established the importance of miRNAs in cancer, leading to an exponential growth in miRNA cancer research. Numerous studies reporting miRNA profiling of cancer specimens were published with the intent to determine the functional significance of individual miRNAs in addition to the speculation that miRNA profiles could have clinical practicality for tumor classification.
3.5.1 Breast Cancer Subtype miRNA Profiles
Early and recent miRNA profiles showed a subset of miRNAs differentially expressed in normal tissues compared to solid tumors including breast cancers. Furthermore, some of these miRNAs were associated with biopathologic features of breast cancer such as ER+/−, PR+/−, cancer stage, nodal status, vascular invasion, proliferation index, and p53 status [118–122]. miRNA profiles were also performed on the different molecular subtypes of breast cancer as defined by gene expression signatures. Furthermore, the use of miRNA profiling was applied to determining a miRNA signature of different mammary cell types [123–128].
Blenkiron et al. showed that 133 miRNAs are differentially expressed in the human breast and in the different molecular subtypes of breast cancer. They first classified 93 cases of primary human breast tumors by gene expression profiling into luminal A, luminal B, basal-like, HER2+, normal-like, and ER+/−. Using a bead-based flow cytometric miRNA expression profiling method, they identified differentially expressed miRNAs in the various molecular subtypes. These miRNAs include pathologically important miRNAs: members within the let-7, miR-200, and miR-10 family; the miR-17–92 and miR-214 clusters; and individual miRNAs such as miR-155, miR-21. Furthermore, they showed miR-100, miR-99a, miR-130a, miR-126*, miR-136, miR-146b, miR-15b, miR-107, and miR-103 can differentiate between the luminal A and luminal B subtypes, in addition to providing a profile for differentiating ER+/− and tumor grade [128].
Sempere et al. used locked nucleic acid array to profile breast cancer cell lines in addition to normal and tumor breast tissue specimens. Then, they implemented an in situ hybridization method to determine the localization of miRNAs within specific cell types of formalin-fixed, paraffin-embedded tissue specimens. The rational was that the miRNAs of interest should be aberrantly expressed within malignant cells rather than the involved stroma, infiltrating lymphocytes, and/or vasculature. They found miR-145 and miR-205 expression in myoepithelial cells of normal mammary epithelial structures, whereas their expression was significantly reduced or eliminated in matching tumor specimens. Let-7a, miR-21, miR-141, and miR-214 were detectable within luminal epithelial cells in normal tissue where let-7a was often decreased while miR-21 was increased in malignant cells. Interestingly, although the authors did not extrapolate on this finding, their heat map indicated elevated expression levels of miR-221, miR-222, miR-214, and miR-21 in ER- tumors [124].
3.5.2 MicroRNA-155
One miRNA rising to prominence in breast cancer is microRNA-155 (miR-155). It was originally identified to be differentially expressed in miRNA profiles [118, 119, 128]. Kong et al. discovered miR-155 may have implications in aggressive, invasive breast cancer using TGFβ induced epithelial to mesenchymal transition in normal mouse mammary gland epithelial cells (NMuMG), an in vitro cell line model often used to study mechanisms of breast cancer metastasis. NMuMG expressing miR-155 exhibits greater invasion and migration, whereas NMuMG without miR-155 had the opposite effect. miR-155 expression also induced cellular plasticity that could be rescued with RhoA reconstitution [129]. In another study, Kong et al. showed miR-155 induced breast cancer cell proliferation and chemoresistance by targeting FOXO3a [130]. Furthermore, Blenkiron et al. showed that miR-155 expression is highly elevated in basal-like and high-grade breast cancer [128].
Chang et al. recently published data showing that the tumor suppressor BRCA1 epigenetically controls miR-155 expression. They demonstrated that the R1699Q moderate risk variant, which exhibits a point mutation in the BRCT domain, loses its ability to repress miR-155, explaining how BRCA1-deficient cells express high levels of miR-155. Orthotopic transplantation of BRCA1+ MDA-MB-468 cells ectopically expressing miR-155 into athymic nude mice exhibited accelerated tumor growth relative to control. Analyses of BRCA1-mutant human breast tumors show inverse correlation between BRCA1 and miR-155 expression [131]. The complementation of in vitro experiments by Kong et al., with the clinical breast cancer profile work by Blenkiron et al., suggested that miR-155 has important clinical implications in aggressive breast cancer. The data of Chang et al. solidified the physiological importance of miR-155 in the critical BRCA1 mutant breast cancers. This suggests miR-155 could be a potential biomarker in addition to a therapeutic target for specific types of aggressive breast cancers [128–131].
3.6 Conclusions
Diagnosis of breast cancer has evolved from observing the physical appearance of the tumor under the microscope to quantifying expression of genes at the molecular level. Although the latter method may seem less subjective and more quantitative, it is still subject to variations including but not limited to sample handling and reagent lot variations among other factors. Implementation should probably not replace the former method supported by the expertise of a trained pathologist. Molecular diagnosis should probably be used in a combinatorial manner, although the cost of doing so might not justify its implementation. Furthermore, basic research in science now covers a variety of subjects such as epigenetics, regulation by non-coding RNAs, identification of cancer stem cells and understanding their functions from genetics to biology, and a vast variety of other disciplines in between. Furthermore, next-generation sequencing technologies and the technological means to integrate and analyze diverse data types on a large scale, which allows investigation of biological networks in diseased state, will likely drive further changes in molecular diagnosis of not only breast cancers but other diseases.
References
1.
Azzopardi JG, Ahmed A, Millis RR (1979) Problems in breast pathology. Major Probl Pathol 11:1–466, i-xvi
2.
Tavassoli FA (1992) Classification of metaplastic carcinomas of the breast. Pathol Annu 27(Pt 2):89–119PubMed
4.
Weigelt B, Reis-Filho JS (2009) Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol 6:718–730PubMed
5.
Veltmaat JM, Mailleux AA, Thiery JP, Bellusci S (2003) Mouse embryonic mammogenesis as a model for the molecular regulation of pattern formation. Differentiation 71:1–17PubMed
6.
Tiede B, Kang Y (2011) From milk to malignancy: the role of mammary stem cells in development, pregnancy and breast cancer. Cell Res 21:245–257PubMed
7.
Woodward WA, Chen MS, Behbod F, Rosen JM (2005) On mammary stem cells. J Cell Sci 118:3585–3594PubMed
8.
Reis-Filho JS, Lakhani SR (2008) Breast cancer special types: why bother? J Pathol 216:394–398PubMed
9.
Simpson PT, Reis-Filho JS, Gale T, Lakhani SR (2005) Molecular evolution of breast cancer. J Pathol 205:248–254PubMed
10.
Vargo-Gogola T, Rosen JM (2007) Modelling breast cancer: one size does not fit all. Nat Rev Cancer 7:659–672PubMed
11.
Visvader JE (2009) Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev 23:2563–2577PubMed
12.
Petersen OW, Polyak K (2010) Stem cells in the human breast. Cold Spring Harb Perspect Biol 2(5):a003160PubMed
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree