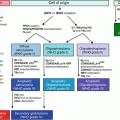
Fig. 17.1
The diagnostic algorithm for lymphoid malignancies commonly accepted in daily practice
17.2.1 Molecular Methods Commonly Used in Lymphoid Malignancies
17.2.1.1 Standard Chromosomal Analysis
Conventional karyotyping is one of standard chromosomal analyses that have been widely used in diagnosis of myeloid leukemias as well as lymphoid neoplasms. Discovery of the recurrent cytogenetic abnormality in Burkitt lymphoma with t(8; 14) and chronic myelogenous leukemia with t(9;22) in early 1960s. [13, 14], is followed by many chromosomal abnormalities described in lymphomas and leukemias. Up to date, certain precursor B lymphoid neoplasms, vast majority of the Burkitt lymphoma, mantle cell lymphoma, follicular lymphoma and a subset of marginal zone lymphomas are associated with recurrent translocations (Table 17.1) [1–3, 15, 16]. Diffuse large B cell lymphomas (DLBCL), can associate with single or multiple breakpoints and translocations, namely BCL6 at 3q and BCl-2 at 18q, “double or triple” hit lymphomas commonly involving MYC and BCL2 and less frequently BCL6 genes [17–20]. Nevertheless, only a minority of T cell lymphomas exhibit recurrent chromosomal translocations. Anaplastic large T cell lymphoma is a good example, often associated with t(2;5) involving NPM1-ALK gene fusion [21], though the other ALK partner genes also encountered.
Most cytogenetic abnormalities seen in lymphomas and leukemias are often detectable by conventional karyotyping by G-banding. Although it is considered to be the “gold standard” technique, some limitations prevent a wide use of this test, including the need to obtain a large volume of fresh tissue and rapid processing. Another problem encountered in cytogenetic testing is that the analysis requires high level of technical skills in culturing tissue, preparing the metaphase cells and analyzing complex karyograms which makes the method labor-intensive, expensive and time consuming [15].
17.2.1.2 Fluorescence In Situ Hybridization (FISH)
Fluorescence in situ hybridization (FISH) is widely used as an ancillary method in diagnosis of lymphomas and leukemias. It is applicable to cytospin preparations from fresh harvested tissue as well as formalin-fixed and paraffin-embedded (FFPE) tissue without requiring tissue culturing. Thus, FISH is a less complex and rapid molecular technique with shorter turnaround time. The main limitation of FISH is the probe specific testing only for specific abnormalities with no genome-wide analysis.
17.2.1.3 Polymerase Chain Reaction (PCR), Reverse Transcription-PCR (RT-PCR)
PCR tests are widely used in lymphomas and lymphoblastic leukemias for clonality analysis of the immunoglobulin genes and T cell receptors (TCR). The testing can be performed on either fresh tissues or FFPE materials. DNA or RNA materials are extracted from submitted specimen according to the study design. Quantitative RT-PCR is also very useful in detecting of the chromosomal translocations that lead to the production of fusion mRNA transcripts. A good example of this fusion transcript and protein generation is cyclin D1-IgH resulting in mantle cell lymphoma [22]. An advantage of PCR in lymphoma and leukemias is that it enables to amplify the fusion transcript; therefore it is superior in detection of minimal residual disease (MRD). PCR analysis for clonality is particularly advantageous in certain circumstances; (1) if the tissue is limited (e.g. small needle biopsies or fine needle aspirations) for immunohistochemical analysis and/or flow cytometry cannot be performed (e.g. FFPE tissue); (2) some B cell lymphoma cells may lack surface light chain antigens so that clonality may be equivalent by flow cytometry. (3) Phenotypic clonality evaluation has not been established for T cell neoplasms, therefore, gene rearrangement studies are helpful determining T cell clonality; (4) differentiation of reactive hematogone response vs. residual/recurrent precursor lymphoid neoplasms with increased immature lymphoblasts [23–26]. On the other hand, there are some limitations when using PCR for diagnostic purposes. False positive gene rearrangement results can be observed especially when testing a small biopsy with small number of lymphocytes as well as various reactive and inflammatory conditions [27, 28]. Furthermore, some T cell lymphomas may have false immunoglobulin (IG) gene re-arrangement [28]. False negative results can also be seen mostly due to technical or biologic factors [29].
17.2.1.4 Single Nucleotide Polymorphism (SNP) Array
SNP array is the most recently introduced molecular method that can analyze the whole genome with higher resolution than the other techniques. An important advantage of SNP array is the ability to detect not only copy number changes but also the regions with loss of heterozygosity without DNA loss or gain. This indicates that the both alleles are homozygous and derived from one parent, “uniparental disomy” (UDP), which is important in certain lymphomas [30–32]. Regions of UDP may harbor one or more genes with inactivating mutations (classical tumor suppressor genes) and since both alleles will have the mutation, the Knudson two-hit rule is fulfilled [15]. The main problem with SNP array is the difficulty in distinguishing acquired abnormalities from polymorphism which may be resolved by comparing to the germline DNA from the same patient [33]. SNP is so far not the replacement of cytogenetics and FISH laboratory, however, it is usually ordered for those patients with normal karyotyping or FISH results when seeking a possible disease related genetic findings.
17.3 Precursor Lymphoid Neoplasms
Precursor lymphoid neoplasms, also termed acute lymphoblastic leukemias (ALL), are one of the most common hematopoietic malignancies affecting pediatric population and a major cause of co-morbidity in adult. The current WHO classification of ALL contains three main subgroups: (1) B-lymphoblastic leukemia/lymphoma (B-ALL), not otherwise specified, (2) B-ALL with recurrent cytogenetic abnormalities, and (3) T-lymphoblastic leukemia/lymphoma (T-ALL) [1] (Table 17.1). ALL is a greatly heterogeneous group of disorders with variety of associated genetic defects that have been defined recently or still unknown. The classical algorithm used in diagnosis of ALL includes a careful morphological examination of peripheral smear and/or bone marrow samples as well as involved tissue with appropriate phenotypic studies supported by relevant testing for karyotype analysis and molecular genetics. Cytogenetic abnormalities are seen in 60–80 % of ALL including recurrent genetic abnormalities.
Table 17.1
The WHO classification of precursor lymphoid neoplasms
I. Precursor lymphoid neoplasms |
|---|
B lymphoblastic leukemia/lymphoma, not otherwise specified (NOS) |
B lymphoblastic leukemia/lymphoma, with recurrent genetic abnormalities |
B lymphoblastic leukemia/lymphoma with t(9;22)(q34;q11.2); BCR-ABL |
B lymphoblastic leukemia/lymphoma with t(v;11q23); MLL rearranged |
B lymphoblastic leukemia/lymphoma with t(12;21)(p13;q22); TEL-AML-1 (ETV6-RUNX1) |
B lymphoblastic leukemia/lymphoma with hyperdiploidy |
B lymphoblastic leukemia/lymphoma with hypodiploidy (Hypodiploid ALL) |
B lymphoblastic leukemia/lymphoma with t(5;14)(q31;q32); IL3-IGH |
B lymphoblastic leukemia/lymphoma with t(1;19)(q23;p13.3); E2A-PBX1(TCF3-PBX1) |
T lymphoblastic leukemia/lymphoma |
17.3.1 B Lymphoblastic Leukemia/Lymphoma with Recurrent Genetic Abnormalities
This is a group of disease characterized by recurrent genetic abnormalities including chromosomal abnormalities and balanced translocations. The distinction of this B-ALL from other types is especially important as they have specific clinical and phenotypic features leading to important prognostic implications.
17.3.1.1 B Lymphoblastic Leukemia/Lymphoma with t(9;22)(q34;q11.2);BCR-ABL1
The Philadelphia (Ph) chromosome is the most common associated genetic anomaly in adult B-ALL, accounting for about 20–30 %. Only about 2–4 % of childhood leukemias harbor Ph chromosome. The lympho blasts have the translocation between the BCR gene on chromosome 22 and the ABL oncogene on chromosome 9 that creates the Ph chromosome. While the ABL breakpoint occurs in the same region (between exons a1 and a2) on chromosome 9, the BCR breakpoints on chromosome 22 can occur in two different regions. The m-bcr creates a fusion protein of 190 kDa (p190) and M-bcr results in a fusion protein of 210 kDa (p210) [34, 35]. The “minor” breakpoint cluster region (m-bcr) between exons 1 and 2 is present in vast majority of childhood Ph + ALL while a “major” breakpoint cluster region (M-bcr) spanning exons 12 to 16 is more common in adult Ph + ALL. The m-bcr creates a fusion protein of 190 kDa (p190) and M-bcr results in a fusion protein of 210 kDa (p210) [34, 35]. Phenotypically, Ph + B-ALL is typically CD10+, CD19+ and TdT+. The expression of myeloid antigens CD13 and CD33 is frequent. CD25 is highly associated with adult Ph + ALL [36].
Cytogenetics is still commonly used in detection of Ph chromosome. In cases of failed or normal cytogenetic results, FISH can be used as an alternative method. Quantitative RT-PCR is a rapid, accurate and sensitive method for the detection of BCR/ABL fusion transcripts. The advantage of test is to quantitatively track the copy number change of transcript before and after therapy. One major disadvantage is that this approach is highly specific and cooperating abnormalities cannot be simultaneously identified [37]. The presence of t(9;22) is a poorer prognostic factor in both children and adults. Combination of tyrosine kinase inhibitor (TKI) with established antileukemic agents is promising in management of these patients.
17.3.1.2 B Lymphoblastic Leukemia/Lymphoma with t(v;11q23); MLL Rearranged
This is a group of acute leukemias harboring translocation between MLL gene at band 11q23 and any of the fusion partners. MLL translocations generate a new chimeric gene, in which the NH2-terminal portion of MLL is fused to the COOH-terminal sequence from multiple different partners [38]. The common result of many of these rearrangements is the expression of a DNA-binding protein that recruits additional histone methyltransferases such as DOT1L and leads to ectopic histone H3 lysine 79-dimethylation [39, 40].
These lymphomas exhibit unique clinical and biologic feature. MLL translocation occurs up to 70 % of infant B-ALL and less frequently in older patients. Typically patients present with very high white blood cell counts and often central nervous system (CNS) involvement at diagnosis. A characteristic feature of ALL with MLL translocations, mostly t(4;11) ALL, is the absence of CD10 in pro-B blasts often expressing myeloid antigen CD15. The expression of chondroitin sulfate proteo-glycan neural-glial antigen 2 (NG2) is also typical in these leukemias. Many fusion genes have been described in ALL with MLL translocation. t(4;11)(q21;q23)/MLL-AFF1 (previously known as MLL-AF4) comprise the largest group in ALL with MLL. Other common partner genes include ENL (19p13) and AF9 (9p22). FLT3 overexpression is also found in the presence of MLL translocations [41]. MLL-AFF1 fusion is easily detectable by RT-PCR or cytogenetics. However, a more complex molecular analysis may be advantageous in detecting other common fusion partners. A dual color break-apart probe may provide detection of all chromosomal abnormalities involved in ALL with MLL translocations by FISH. Leukemias with MLL gene rearrangements usually have poor prognosis. The specific type B-ALL is also associated with adverse outcome, particularly when it occurred in infant <6 months.
17.3.1.3 B Lymphoblastic Leukemia/Lymphoma with t(12;21)(p13;q22); TEL-AML1 (ETV6-RUNX1)
This is the most common leukemia in childhood, accounting 25 % of all B-ALLs with a very favorable prognosis. It is not identified in infants and barely found in adults. It has similar clinical and phenotypic features to other types of ALL. CD13 myeloid antigen expression is common with no indication of mixed phenotype acute leukemia. t(12;21) results in the production of TEL-AML1(ETV6-RUNX1), a fusion protein with important role in leukomogenesis [42]. The product protein acts in a dominant negative fashion to interfere with normal function of the transcription factor RUNX1. It has been shown that the translocation occurs early in life, prenatally, as “preleukemic clone” but leukemia develops many years later [43]. There is evidence that secondary changes are needed for overt leukemia including frequent additional changes include losses of the EBF1, ETV6 and PAX5 genes and gains of chromosome 21 and chromosome arm Xq [44]. The fusion protein can be detected by RT-PCR. As t(12;21)(p13;q22) is invisible at cytogenetic level, FISH is usually used to detect the translocation applying an extra signal or dual color fusion probe. Furthermore, detecting deletion of second ETV6 allele and additional fusion signals is also important in this type of B-ALL, possibly leading a longer survival [45]. FISH provides information on the status of second ETV6 homologue.
17.3.1.4 B Lymphoblastic Leukemia/Lymphoma with Hyperdiploidy
Hyperdiploid B-ALL is characterized by blasts containing 51–65 chromosomes, in the absence of translocations or other structural alterations. This subtype accounts for 25 % of B-ALL. It is common in children and has a good prognosis with a survival rate up to 80 % at 5 years. Hyperdiploid B-ALL has no unique features to distinguish it from other subtypes. Hyperdiploid is characterized by gain of chromosomes, typically + X, +4, +6, +10, +14,+17, +18, and +21. The best method for the detection of chromosomal gain in hyperdiploid B-ALL is the standard karyotyping. Cytogenetics may fail in a portion of cases [46, 47]. FISH with centromeric probes specific for the trisomies of chromosomes 4, 10, 17, and 18 may be helpful to identify hidden hyperdiploidy [48, 49]. Identified trisomy 4, 10, and 17 link to a good prognosis. Flow cytometry to measure DNA index analysis can also be used.
17.3.1.5 B Lymphoblastic Leukemia/Lymphoma with Hypodiploidy (Hypodiploid ALL)
Hypodiploid B-ALL is characterized by blasts containing < 44–45 chromosomes including near-haploid ones (23–29 chromosomes). It is seen in both children and adults, although near-haploid ones are more common in children. This subgroup is associated with a worse prognosis. The overall clinical, morphologic and phenotypic features are not distinguishable from other B-ALL. By definition, there is loss of from single or more chromosomes to near haploid with or without structural abnormalities in the remaining chromosomes. The structural anomalies including translocations are more common in the cases with chromosome number between 30 and 40. Detection of hypodiploidy in this group could be problematic. One characteristic feature is the gain of some specific chromosomes onto the haploid chromosomes and the presence of a population of cells with an exact doubling of this chromosome number [50]. This may result in a near diploid or even hyperdiploid karyotype. In such cases, standard karyotyping may dismiss the hypodiploid line. Flow cytometry can be used to detect hypodiploidy in these cases with the advantage of more rapid processing. Flow cytometry is also not affected by mitotic index of the cell population. However, flow cytometry does not identify the specific chromosomes with numeric changes, nor does show the additional structural anomalies which may have potential clinical and biologic importance [51]. Therefore, it is useful to use both techniques in tandem. FISH may also detect certain chromosomes with hypodiploidy; however, FISH detection may be difficult when near-haploid or doubled populations present.
In summary, hypodiploidy and near-haploidy in B-ALL carries a prognostic significance and molecular testing should be carefully applied in detecting chromosomal loss when this subgroup is clinically suspected.
17.3.1.6 B Lymphoblastic Leukemia/Lymphoma with t(5;14)(q31;q32);IL3-IGH
This subgroup is very rare and accounts for < 1 % of all ALL. It has been reported in all age groups. The eosinophilia is reactive and not part of the neoplastic process. The features and prognosis are otherwise similar to other B-ALL. The blast count has not shown to be a prognostic factor in this type of ALL. Patients typically have variable circulating eosinophilia due to overexpression of the IL3 gene following the functional rearrangement between IL3 gene and IGH@ gene. The translocation may be detected using standard karyotyping, RT-PCR and/or less commonly FISH.
17.3.1.7 B Lymphoblastic Leukemia/Lymphoma with t(1;19)(q23;p13.3);E2A-PBX1 (TCF3-PBX1)
ALL with t(1;19) is fairly common in children, accounting for 6 % of all ALL. Clinical features are similar to those of other ALL patients. Although they used to be classified in the poor prognosis group, with therapeutic advances, they are regarded as standard risk [52, 53]. Although they do not differ morphologically from other types, blasts in this group are pre-B type and typically express CD19, CD10 and cytoplasmic μ (cμ) with less or no CD34. Strong CD9 expression is also characteristic. TCF3 gene encodes two transcriptional proteins E12 and E47 which play a critical role in B-cell maturation while PBX1 is a homeobox gene not expressed in B or T-cells [54]. Translocation between TCF3 (previously E2A) gene, on chromosome 19, and PBX1 gene, on chromosome 1, results in production of a fusion protein that has an oncogenic role. Recently, a variant subtype harboring translocation between TCF3 and HLF genes related to a worse prognosis. Cytogenetics and FISH as well as RT-PCR are useful to detect the associated translocations in majority of the cases. The diagnosis of this subtype warrants the careful evaluation by both phenotypic features and translocation status, as an identical t(1;19) can be seen in a portion of B-ALL not necessarily associated with TCF3-PBX1 translocation.
17.3.1.8 Other Cytogenetic Abnormalities Detected in B-ALL
Up to 50 % of all B-ALL show abnormalities involving genes associated with B cell development including PAX5, TCF3,EBF1, LEF1, IKZF1 and IKZF3 and cell cycle controlling genes such as CDKN2A, CDKN1B and RB1 [55–59]. A cryptic translocation involving sex chromosomes, t(X;14)(p22;q32) or t(Y;14)(p11;q32), involving IGH@ and CRLF2 genes, and deletion within PAR1, resulting in P2RY8-CRLF2 fusion has been reported [60–62]; possibly activating JAK-STAT signaling pathway and associated with a worse prognosis [63–65]. Intrachromosomal amplification of chromosome 21 (iAMP21) is a distinct chromosomal abnormality in B-ALL and associated with adverse prognosis and poor response to therapy [66]. Despite the variability between patients in the morphology of the abnormal chromosome 21, they consistently show multiple, extra copies of the RUNX1 (AML1) gene, tandemly repeated along the length of the abnormal chromosome [67]. Genomic and expression analysis has further characterized this abnormality, demonstrating a common region of amplification (CRA) between 33.192 and 39.796 Mb on chromosome 21 (which includes RUNX1) [68]. FISH can also be used to detect this abnormality.
17.3.1.9 B Lymphoblastic Leukemia/Lymphoma, NOS
Diagnosis of B-lymphoblastic leukemia/lymphoma (B-ALL), NOS is based on clinical, morphological and phenotypic findings when the aforementioned cytogenetic abnormalities are not indentified. Gene re-arrangement by PCR is a commonly used as a supportive diagnostic tool and also is also helpful in determining the minimal residual disease in some settings. Studies have showed that nearly all cases have clonal DJ rearrangements of the IGH@ gene along with T cell gene rearrangement (TCR) in a majority of the cases (up to 70 %) [30, 69]. The B cell clonality in B-ALL can be detected by PCR amplification of the IG and/or TCR VDJ region. The secondary change of TCR gene in B-ALL is considered reactive with uncertainty in pathophysiology, being worthy of further investigation. SNP might be an alternative way to detect any underlying genetic aberration if it is applicable in clinical laboratory.
17.3.2 T Lymphoblastic Leukemia/Lymphoma
T lymphoblastic leukemia/lymphoma is a neoplasm of precursor T cells which are typically medium to large sized blasts with dispersed chromatin, inconspicuous nucleoli and scant cytoplasm. It involves bone marrow and blood (T-ALL) or lymphoid organs or extranodal sites (T-LBL). Both forms are more common in children than adults. The blasts usually express TdT with often T cell antigens CD2, cytoplasmic CD3, CD4, CD5, CD7, and CD8 and CD1a. Aberrant expression of myeloid markers (CD13 and CD33) can be seen in up to 30 % of the cases [70]. Coexpression of CD117 occurs less frequently but often associated with FLT3 mutation [71]. Multiple genetic aberrations encounter in T-ALL, but none of them is T-ALL specific. Translocations involving TCR loci are the most common cytogenetic anomalies observed in T-ALL/LBL. t(7;10)(q34;q24) and t(10;14)(q24;q11) are most common in adults involving HOX11(TLX1) gene whereas t(5;14)(q35;q32) involving HOX11L2 (TLX3) gene is commonly found in children [6]. As reported, dysregulation of HOX-type transcription factors is found in 30–40 % of cases of T-ALL [72]. Characteristically, HOX-type T-ALLs often accompany with activating NOTCH1 mutations and CDKN2A loss of function, NUP214-ABL amplifications, C-MYB tandem duplications, PHF6 mutations, PTPN2 deletions, and WT1 mutations [6, 72–78], which are not present in non-HOX-type T-ALL. However, the role of these genetic changes in leukemogenesis is not well understood. The unique genetic features are not only helpful in view of diagnosis but also guide future clinical therapy. An in vitro study has demonstrated that a blockade of both TLX1/HOX and NOTCH could inhibit cell cycle progression in TLX1 T-ALL cell line 9,490 cells (Lesley A. 2011). Mutations that may occur in T-ALL/LBL are important. Mutation of NOTCH oncogene is the most common mutation and is seen about 60 % of cases. NOTCH proteins are transmembrane receptors and are important regulators of T cell differentiation and activation. Recent studies showed that MYC is upregulated by activated NOTCH1 in T-ALL which stimulates the growth of T-ALL cells [46, 79]. N-RAS and FLT3 are other mutation seen in T-ALL [80, 81]. Another common rearrangement is the formation of fusion genes, mostly in children. SIL/TAL1 fusion gene results from a cryptic deletion of chromosome 1 and is seen in up to 30 % of childhood T-ALL. Important translocations resulting in fusion gene formation includes t(10;11) involving CALM/AF10 genes in 10 % of all cases and less frequently t(11;19) involving MLL gene. Other MLL gene translocations {t(6;11), t(10;11), t(X;11) and t(4;11)} seen in some T-ALL cases [6]. Philadelphia chromosome has also been reported in rare cases [82]. Deletions may be detected in as many as 30 % cases. 9p21 involving p16 gene and del(6q) are the important ones. Almost all cases show clonality in T cell receptors with up to 20 % accompanying IGH@ gene rearrangement [83].
17.4 Mature Lymphoid Neoplasms
Lymphoid neoplasms are clonal tumors with malignant transformation of normal lymphoid cells at various stages of differentiation and comprise the sixth most common group of malignancies worldwide in men and women. Although certain infectious agents, autoimmune disorders and immunosuppression-related conditions have been linked to some lymphomas, the etiology in the majority of lymphomas remains unclear. Morphology and immunophenotype are generally sufficient to make the diagnosis of most lymphomas. Genetic features have begun to play an important role in classification and diagnosis of mature lymphomas. The recent advances in molecular genetics have enlightened lymphoma biology and provide useful testing that can be integrated with the conventional diagnostic methods widely used in hematopathology. Although isolated use of molecular tests is not of any help in lymphoma diagnosis, the use of these tests has clear utility to facilitate the diagnosis, in particularly morphologically and phenotypically challenging cases. Also some molecular markers have prognostic implications in certain lymphomas. Mature B and T/NK cell lymphomas with recurrent cytogenetic abnormalities are discussed below.
17.4.1 Mature B Cell Neoplasms
Mature B cell lymphomas comprise over 90 % of lymphoid neoplasms worldwide. They are common in Western world and account for 4 % of new cancers each year [1]. The most common types are diffuse large B cell lymphoma and follicular lymphoma. The majority of the B cell lymphomas recapitulate the stages of normal B cell proliferation, except for some types, e.g. hairy cell leukemia, that do not correspond to any maturation stage (Table17.2). Diagnosis of B cell lymphomas, like other mature lymphoid neoplasms, is usually based on histomorphologic and phenotypic features in conjunction with genetic and molecular profiles.
Table 17.2
The WHO classification of mature B cell neoplasms
II. Mature B cell neoplasms |
|---|
Chronic lymphocytic leukemia/small lymphocytic lymphoma |
B-cell prolymphocytic leukemia |
Splenic marginal zone lymphoma |
Hairy cell leukemia |
Splenic B-cell lymphoma/leukemia, unclassifiable |
Splenic diffuse red pulp small B-cell lymphoma |
Hairy cell leukemia-variant |
Lymphoplasmacytic lymphoma |
Heavy chain disease |
Gamma heavy chain disease |
Mμ heavy chain disease |
Alpha heavy chain disease |
Plasma cell neoplasms |
Monoclonal gammopathy of undetermined significance (MGUS) |
Plasma cell myeloma |
Solitary plasmacytoma of bone |
Extraosseous plasmacytoma |
Monoclonal immunoglobulin deposition diseases |
Extranodal marginal zone lymphoma of mucosa-associated lymphoid tissue (MALT lymphoma) |
Nodal marginal zone lymphoma |
Follicular lymphoma |
Primary cutaneous follicle center lymphoma |
Mantle cell lymphoma |
Diffuse large B-cell lymphoma (DLBCL), NOS |
T cell/histiocyte-rich large B-cell lymphoma |
Primary DLBCL of the CNS |
Primary cutaneous DLBCL, leg type |
EBV positive DLBCL of the elderly |
DLBCL associated with chronic inflammation |
Lymphomatoid granulomatosis |
Primary mediastinal (thymic) large B-cell lymphoma |
Intravascular large B-cell lymphoma |
ALK positive large B-cell lymphoma |
Plasmablastic lymphoma |
Large B-cell lymphoma arising in HHV8-associated multicentric Castleman disease |
Primary effusion lymphoma |
Burkitt lymphoma |
B-cell lymphoma, unclassifiable, with features intermediate between DLBCL and Burkitt lymphoma |
B-cell lymphoma, unclassifiable, with features intermediate between DLBCL and Hodgkin lymphoma |
17.4.1.1 Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma
Chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) is a neoplasm of monomorphic small B cells with a fairly unique phenotype. CLL is the most common leukemia in the Western world affecting mainly the elderly population. The neoplastic cells often coexpress CD5 and CD23 along with characteristically dim CD20, CD79b and dim surface immunoglobulin expression. CLL is the term used when long standing lymphocytosis (≥ 5 × 109/L monoclonal lymphocytes for at least 3 months) is present in peripheral blood (PB) with or without bone marrow (BM) involvement [1]. The term SLL refers to the less frequent form with same phenotype to CLL but non-leukemic with the tissue morphology. The diagnosis of SLL requires lymphadenopathy, no cytopenias due to BM involvement by CLL/SLL and <5 × 109/L peripheral blood B cells [1].
CLL is a heterogenous disease with variable clinical outcome in the light of variety of genetic abnormalities. Importantly, CLL has the highest familial genetic predisposition in all hematologic malignancies. A polymorphism in the death-associated protein kinase 1 (DAPK1) gene was associated with familial CLL in a single family, but this allele has not been found again in a larger number of cases [84]. The involved genes in genetically predisposed CLL are still yet to be defined. Approximately 80 % of cases have cytogenetic abnormalities. Although some of these abnormalities can be detected in about 40 % of cases by standard karyotyping, FISH is essential in evaluation [85]. The FISH CLL panel including probe CCND1 study is used to exclude mantle cell lymphoma and predict the prognosis of CLL. The most common recurrent abnormality seen in CLL is deletion of 13q14, approximately 55 % of cases, which is also associated with the best prognosis [86]. Twenty percentage of cases have deletion of 11q22-23which corresponds to loss of ataxia telangiectasia mutated (ATM) gene. This abnormality is associated with bulky lymphadenopathy and poor clinical outcome [87]. Deletion of 17p13 is seen in ~7 % of cases majority of which is related to loss or mutation of TP53 tumor suppressor gene. Deletion 17p13 is often leads a poor prognosis with chemotherapy, particularly Fludarabine resistance in these patients [88–90]. TP53 mutation with or without del(17p13) is associated with a worse prognosis [91, 92] CLL with trisomy 12 often has an atypical morphology and bright aberrant expression of CD11c on the neoplastic B-cells. FISH is sufficient in detecting the chromosomal anomalies but not gene mutations A novel molecular technique, Multiplex Ligation-dependent Probe Amplification (MLPA), can be used to complement FISH [16].
Status of immunoglobulin variable domain genes, Ig VH gene, in remains the “gold standard” prognostic feature in CLL patients. Presence of IGH variable domain gene somatic hypermutation (SHM) is seen 50–60 % of cases; whereas unmutated cases comprise 40–50 % with a worse prognosis [93]. Despite the prognostic importance, the test cannot be performed widely due to its complexity. In this case, the mutation status can be analyzed through a surrogate marker, such as ZAP-70 expression using flow cytometry or infrequently immunohistochemistry. The expression of ZAP-70 corresponds to the absence of IGH SHM in the vast majority of the cases [94, 95]. Recent studies also demonstrated NOTCH1 activating mutations associated with worse prognosis in CLL [96].
17.4.1.2 B Cell Prolymphocytic Leukemia
B cell prolymphocytic leukemia (B-PLL) is the neoplasm of prolymphocytes involving peripheral blood, bone marrow and often spleen. Diagnosis requires prolymphocytes exceeding 55 % of the lymphocytes in the blood. It can be transformed from CLL. The typical morphology of prolymphocytes includes medium size with round nuclei and a prominent central nucleolus. Neoplastic cells strongly express surface immunoglobin, along with B cell antigens (CD19 and CD20). B-cells show often loss of CD23. B-cells with CD5 and CD23 co-expression are seen about 20 % of cases. Variable FMC-7 expression is noted. ZAP-70 and CD38 are expressed in half of the cases with no correspondence to IGH mutation status [97].
Immunoglobulin genes are clonally rearranged in half of the cases. Deletion of 17p and related TP53 mutation has been demonstrated in 50 % of cases associated with poor prognosis and therapy resistant disease [98, 99]. Some case may also have 13q14 by FISH [97].
Initially, t(11;14)(q13;q32) had been demonstrated in “B-PLL”; however these cases are now considered leukemic forms of mantle cell lymphoma [100].
17.4.1.3 Splenic Marginal Zone Lymphoma
According to the WHO classification, splenic marginal zone lymphoma (SMZL) is neoplasm of small B lymphocytes which expansion of splenic white pulp, efface the follicle mantle zone and emerge with marginal zone of larger cells; both small and large cells invade the red pulp [1]. Splenic hilar lymph nodes and bone marrow are usually involved as well as peripheral blood by circulating lymphoma cells with villous projections, usually with polar orientation. Lymphoma cells express surface IgM and often IgD along with B cell markers. Annexin A1 and cyclin D1 are negative with rare CD103 expression. Immunoglobulin heavy and light chains are often re-arranged with somatic hypermutation (SHM). Allelic loss of chromosome 7q31-32 has been reported in up to half of the cases [101]. Overexpression of cyclin dependent kinase 6 (CDK6) genes through chromosome 7q21 has been reported in few cases with possible role in lymphomagenesis [102]. BCL2 and t(14;18) are absent. The presence of 7q deletion and unmutated IgVH in SMZL are possibly associated with adverse clinical outcome [103, 104].
17.4.1.4 Lymphoplasmacytic Lymphoma
Lymphoplasmacytic lymphoma (LPL) is characterized by clonal small B cells, plasmacytoid and plasma cells involving bone marrow and less frequently nodes and spleen. The neoplastic cells are typically IgM-secreting and clinically associated with hyperviscosity, autoimmune disorder, and cryoglobulinerimia. Phenotypically, LPL express both B-cell and plasma cell (subset) markers. Immunoglobulin genes are usually clonally arranged with hypermutation. No specific recurrent cytogenetic abnormality has been described in LPL. Deletion of chromosome 6q may be seen but not consistently and minority of patients showed trisomy 3 or 18 [105, 106]. Recently, Treson et al. reported that MYD88 L265P somatic mutation is detected in LPL/Waldenström’s macroglobulinemia, which can be useful in distinguishing LPL from the other B-cell lymphoma with similar morphologic and immunophenotypic features [107]
17.4.1.5 Plasma Cell Myeloma
Plasma cell myeloma is the neoplasm of mature and clonal plasma cells involving bone marrow with widespread symptoms and findings from asymptomatic to prominent end-organ damage. Clinical, histomorphologic and phenotypic findings are often sufficient for a definitive diagnosis. Genetic evaluation is particularly important as for the prognosis and therapy effectiveness. Immunoglobin heavy and light chains are clonally arranged in myeloma patients. Plasma cell myeloma arises from post-germinal center B cells resulting in IGH SHM.
Standard karyotyping may reveal cytogenetic abnormalities in about one third of myelomas [108, 109]. The main difficulty in karyotype analysis of myeloma is the low-yield metaphases generated from myeloma cells due to low proliferation rate. Therefore; FISH is more commonly used in detecting myeloma related cytogenetics and reveals more than 90 % of the abnormalities [110–112]. When able to analyze sufficient sample, the karyotype in these patients is usually complex, mostly hyperdiploidy. Hyperdiploidy has been defined in 40–65 % of myelomas [109, 113]. These gains in the chromosomes are random and often involve chromosomes with odd numbers including 3, 5, 7, 9, 11, 15, 19 and 21 [114]. Other less common type of karyotype abnormality includes structural re-arrangements in the absence of hyperdiploidy. Most of these cases include specific rearrangements involving the IGH gene located at 14q32 and comprise 55–70 % of all cases [115–118]. These rearrangements are typical reciprocal translocations with various chromosomal partners leading to upregulation of the target genes. The most frequent 14q32 partners are 11q13 (involving CCND1 (CyclinD1); 20 % of patients) [119, 120], 4p16 (involving both FGFR3 and MMSET; 15 % of patients) [121, 122], as well as 16q23 and 20q11 (targeting MAF genes; 2–5 % of patients each) [123–125]. Both IGH@ translocations and hyperdiploidy appear to be early events in the oncogenesis in plasma cell neoplasms, unified by upregulation of various cyclin D genes (D1, D2, and/or D3) [126, 127].
Other chromosomal changes seen in myeloma include monosomy 13 which may be seen in about half of the cases considered an early event in the pathogenesis [110, 117, 128]. Additional two secondary events are also recurrent: gains of the long arm of chromosome 1 (1q21), in one third of patients, and del 17p, observed in approximately 10 % of patients which are related to disease progression from MGUS to myeloma as well as disease relapse [129, 130].
Activating mutations of KRAS- or N-RAS may be present in up to 40 % of cases and are likely associated with disease progression [117, 131]. Translocations involving MYC gene and activation of NF-kappa B pathway have also been shown in myeloma patients [132, 133]. Chapman et al. recently showed 4 % BRAF mutation in myeloma patients, a novel discovery in myeloma suggesting therapeutic implication [134]. Recent studies also emphasized the significance of Phosphatidylinositide 3-kinase (PI3K)/protein kinase B (PKB) pathway, also known as the Akt pathway, in myeloma [135]. The PKB/Akt kinase family includes 3 isoforms (PKBα/Akt1, PKBβ/Akt2, and PKBγ/Akt3) that are encoded by separate genes and that are involved in the regulation of apoptosis, proliferation, motility, and energy metabolism. In many tumors including myeloma, activation of Akt is usually indicative of poor prognosis [136].
17.4.1.6 Extranodal Marginal Zone Lymphoma of Mucosa-Associated Lymphoid Tissue (MALT Lymphoma)
This is an often indolent extranodal lymphoma composed of heterogenous small B cells associated with various precursor lesions described, e.g. Helicobacter pylori gastritis and autoimmune diseases. Neoplastic lymphoid cells infiltrate the epithelium causing destruction of normal architecture and are characterized as “lymphoepithelial lesion”. There is several recurrent cytogenetic changes described in MALT lymphoma. The t(11;18)(q21;q21) resulting in fusion of API2 and MALT1 genes is detected in approximately one third of cases by standard karyotyping. RT-PCR or FISH studies are helpful in detecting this defect and can be performed on either fresh cells or PEFF tissue [46]. Importantly, the cases with t(11;18) are resistant to therapy targeting H. pylori [137]. Other described translocations associated with MALT lymphoma include t(1;14)(p22;q32), t(14;18)(q32;q21), (3;14)(p14.1;q32) and t(1;14) (p22;q32), showing fusion of the N-terminus of the API2 gene to the C-terminus of the MALT1 gene or trancriptional gene deregulation in BCL10, MALT1 or FOXP1 [138, 139, and 140]. t(1;14) (p22;q32), involving BCL10 gene is usually seen in MALT lymphoma of lung and stomach [139, 140]. Immunohistochemical evaluation of BCL10 protein nuclear staining can be correlated with the presence of translocation [141]. t(14;18)(q32;q21), resulting fusion of IGH/MALT1, is associated with transcriptional dysregulation of MALT1 and accumulation of cytoplasmic BCL10 [138]. This translocation is most common in parotid, liver and ocular MALT lymphoma [139, 140]. The translocation by karyotyping seems indistinguishable from that seen in follicular lymphoma and an attention should be paid when diagnosing MALT lymphoma. MALT lymphoma arising in thyroid, ocular adnexa and skin can be associated with t(3;14) (p14.1;q32) involving FOXP1 and IGH@ [140, 142]. Trisomy 3, 18, and less commonly other chromosomes may be seen less frequently in MALT lymphomas. BCL6 translocation can also rarely be seen resulting in fusion to the IGH@ gene [143].
17.4.1.7 Follicular Lymphoma
Follicular lymphoma (FL) is a clonal neoplasm of germinal center B cells including both small centrocytes and large centroblasts with a variable morphology and grading at the diagnosis. Approximately 90 % of the cases harbor the characteristic translocation, t(14;18), involving BCL2 and IGH@ genes. That can be detected by cytogenetic analysis or FISH as a more sensitive tool. BCL2 overexpression results in decreased apoptosis in the lymphoma cells. Importantly, a subportion of cases, mostly high grade FL, may lack BCL2 rearrangement [144]. A special attention needs to be paid in the situation in order to avoid a misinterpretation of “reactive germinal center cells”. Morphologic features by devoiding of “tangible body macrophage”, loss of normal polarity and significantly increased centroblasts are diagnostic clues when lack of the specific gene/protein expression. Furthermore, some FL, usually seen in elderly population, may lack BCL2 but harbor BCL6 re-arrangement. These cases have a different phenotype than typical FL; CD10-, BCL2- and BCL6+ as well as MUM1+ [145, 146]. The BCL2 re-arrangement status may vary depending on the localization of primary FL. For instance, primary cutaneous FL is frequently BCL2 negative and often lack IgH-BCL2 fusion product/t(14;18) when analyzed by FISH [147]. Pediatric FL also lacks BCL2 and the translocation.
Other common cytogenetic abnormalities seen in 90 % of FL include 1p-, 6q-, 17p-, +7, +8, +12 and + X [148, 149]. Deletions of 6q25-q27 and 9p21 are associated with poor prognosis [150].
Transformation to a more aggressive lymphoma from FL has been described. Translocations resulting in inactivation of TP53 and p16 and activation of MYC are associated with transformation to a higher grade disease [151–154]. Per literature, the majority of FL transforms into diffuse large B-cell lymphoma (DLBCL). Transformation to Burkitt-type lymphomas or precursor B-cell lymphoblastic lymphoma is also reported in a small subset of FL [155, 156].
17.4.1.8 Mantle Cell lymphoma
Mantle cell lymphoma (MCL) is a neoplasm of small to medium sized B cells with typical CD5 co-expression and (CCND1) translocation. Translocation between CCND1 on chromosome 11 and IGH@ on chromosome 14 leading to overexpression of CCND1 is characteristic and seen in almost all cases. CCND1 gene encodes a cell cycle protein, dysregulation of which by IGH@ gene enhancer region results in possibly overcoming the cell cycle suppressors leading to MCL development [157, 158]. FISH analysis of CCND1 is the preferred and commonly used method in diagnosis of MCL. Rare cases of MCL do not overexpress cyclin D, in which a higher expression of cyclin D2 and cyclin D3 is present and these cases have identical clinical and morphologic features and same genomic profile [159]. A subset of these cases harbor t(2;12)(p12;p13) or t(12;14)(p13;q32) fusing cyclin D2 to kappa light chain (IGK@) or IGH@, respectively. RT-PCR is superior in assessing cyclin D2 translocations. Several other recurrent genetic abnormalities have been demonstrated in MCL, including losses of 9p21.3, 11q22-q23, and 22q11.22, and gains of 10p11.23 and 13q31.3 [160, 161]. Specific mutations and deletions in TP53 (17p13), p16 (CDKN2A), ATM, and CHEK2 have also been frequently noted in MCL [162, 163]. The cases with TP53 are rather in blastoid morphology compared to conventional MCL and associated with poor prognosis. Deletion of 13q14 and trisomy 12 also can be seen in MCL [164]. MCL is often encountered as an aggressive lymphoma by nature. Recent studies demonstrated an indolent variant sharing a common genomic profile with conventional MCL. SOX11 gene, which has a role in cell differentiation, is found to be associated with indolent variant MCL [165, 166]. Immunohistochemical testing is available to evaluate SOX11 protein.
17.4.1.9 Diffuse Large B Cell Lymphoma
Diffuse large B cell lymphoma, not otherwise specified (DLBCL, NOS) is a very heterogenous neoplasm of large B cells with variable localization, morphology and clinical behavior. DLBCL, NOS accounts for 25–30 % of adult non-Hodgkin lymphomas and common in elderly population. It can arise as the primary or as transformation or progression from a lower grade lymphoma and clinically presents a rapidly enlarging mass. A subset of cases may be associated with EBV infection in the absence of immunodeficiency. DLBCL may arise in the nodal or extranodal sites, with gastrointestinal tract being the most common extranodal site. Common morphological variants include centroblastic, immunoblastic, and anaplastic as well as rare variants, e.g. with spindle morphology [1]. By immunohistochemical features, DLBCL, NOS is divided into three groups; (1) CD5-positive DLBCL; (2) germinal center B-cell-like (GCB) and (3) non-germinal center B-cell-like (non-GCB). Molecular subgroups include germinal center B-cell-like (GCB) type and activated B cell-like (ABC) type [1].
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree