Following activation by their cognate ligands, cytokine receptors undergo intracellular routing toward lysosomes, where they are degraded. This review focuses on the signaling function of the G-CSFR in relation to the dynamics of endosomal routing of the G-CSFR. Mechanisms involving receptor lysine ubiquitination and redox-controlled phosphatase activities are discussed. Specific attention is paid to the consequences of G-CSFR mutations, acquired in patients with severe congenital neutropenias who receive G-CSF therapy, particularly in the context of leukemic transformation, a major clinical complication of the disease.
Key points
- •
Although granulocyte colony-stimulating factor (G-CSF) has been routinely used for more than 2 decades to treat various types of severe congenital neutropenia (SCN), the underlying mechanisms of G-CSF hyporesponsiveness or nonresponsiveness have remained largely elusive.
- •
A thorough understanding of the biology of G-CSF receptor (G-CSFR) activation and signaling function in relation to endosomal and lysosomal trafficking will be useful in identifying compounds that may act synergistically with G-CSF to overcome hyporesponsiveness or nonresponsiveness in SCN.
- •
It may shed light on how abnormal function of the G-CSFR, caused by mutations that prevent their normal intracellular trafficking, contribute to a premalignant state of hematopoietic stem and progenitor cells.
- •
Specifically, it remains to be resolved whether G-CSF merely drives the clonal expansion of the “sick” stem cells in SCN or actively contributes to the leukemic process through elevated production of ROS after activation of the truncated G-CSFR, leading to chronic DNA damage, acquisition of additional mutations, and rewiring of signaling and transcription regulatory processes.
Granulocyte colony-stimulating factor and its receptor
Granulocyte colony-stimulating factor (G-CSF) and its receptor (G-CSFR) control neutrophil production under basal circumstances and during episodes of bacterial infections, a condition referred to as “emergency” granulopoiesis. Mice deficient for either G-CSF or G-CSFR suffer from severe neutropenia and are hyper-susceptible to infections, confirming the nonredundant role of the G-CSF/G-CSFR signaling axis in granulopoiesis and host defense. G-CSFR is a single transmembrane protein and a member of the cytokine receptor superfamily. The extracellular part contains an immunoglobulinlike (Ig-like) domain, a cytokine receptor homology domain (CRH domain) comprising a tryptophan serine repeat (WSXWS) and conserved cysteines, and 3 fibronectin type III (FNIII) modules ( Fig. 1 ). G-CSF binds to G-CSFR in a 2:2 ratio, resulting in the homodimerization of receptor proteins required for G-CSFR activation.
Like other members of this superfamily, G-CSFR activates the Janus kinase/signal transducer and activator of transcription (JAK/STAT), phosphoinositide-3 kinase/Akt (PI-3K/Akt) and p21 RAS /mitogen activated protein kinase (MAPK) signaling pathways. In addition, a number of novel mechanisms involved in G-CSF signaling have been proposed. The G-CSFR activates 3 members of the JAK family (JAK1, JAK2, and TYK2) and 2 STAT members (STAT3 and STAT5). The temporal kinetics of STAT3 and STAT5 activation are markedly distinct; whereas STAT3 stays activated on prolonged G-CSFR activation for hours (or even days), activation of STAT5 peaks after 15 minutes and then declines rapidly. The gene encoding the suppressor of cytokine signaling 3 (SOCS3) is the most prominent direct transcriptional target of STAT3 in myeloid progenitors ; G-CSF–induced STAT5 targets in myeloid precursors remain ill-defined.
The cytoplasmic domain of the G-CSFR comprises 4 conserved tyrosine (Y) residues, which in the phosphorylated (p) state serve as binding sites for Src homology 2 (SH2) domain-containing signaling proteins. These pY-coupled pathways are dispensable for G-CSF–induced granulopoiesis but orchestrate the signaling output of the G-CSFR, thereby controlling the proliferation/differentiation balance in neutrophil production under basal and emergency conditions. For instance, pY729 (pY728 in mice) is a recruitment site for the inhibitory SOCS3 protein, whereas the adapter proteins Shc and Grb2, involved in proliferation and prosurvival signaling through activation of p21Ras and PI-3K/Akt, bind to pY764 (pY763 in mice). Details of pY-linked signaling pathways activated by the G-CSFR have been reviewed earlier and are summarized in Fig. 1 . Interestingly, SOCS3 does not only act in a classical negative feedback loop to attenuate G-CSF signaling, but is also involved in a transmodulatory role of the proinflammatory cytokine interferon-gamma (IFNγ) in the control of myelopoiesis. While enhancing monocytic differentiation through activation of the transcription factors IRF8 and PU.1 in myeloid progenitor cells, IFNγ reduces G-CSF–driven neutrophil differentiation via STAT3-mediated induction of SOCS3.
Granulocyte colony-stimulating factor and its receptor
Granulocyte colony-stimulating factor (G-CSF) and its receptor (G-CSFR) control neutrophil production under basal circumstances and during episodes of bacterial infections, a condition referred to as “emergency” granulopoiesis. Mice deficient for either G-CSF or G-CSFR suffer from severe neutropenia and are hyper-susceptible to infections, confirming the nonredundant role of the G-CSF/G-CSFR signaling axis in granulopoiesis and host defense. G-CSFR is a single transmembrane protein and a member of the cytokine receptor superfamily. The extracellular part contains an immunoglobulinlike (Ig-like) domain, a cytokine receptor homology domain (CRH domain) comprising a tryptophan serine repeat (WSXWS) and conserved cysteines, and 3 fibronectin type III (FNIII) modules ( Fig. 1 ). G-CSF binds to G-CSFR in a 2:2 ratio, resulting in the homodimerization of receptor proteins required for G-CSFR activation.
Like other members of this superfamily, G-CSFR activates the Janus kinase/signal transducer and activator of transcription (JAK/STAT), phosphoinositide-3 kinase/Akt (PI-3K/Akt) and p21 RAS /mitogen activated protein kinase (MAPK) signaling pathways. In addition, a number of novel mechanisms involved in G-CSF signaling have been proposed. The G-CSFR activates 3 members of the JAK family (JAK1, JAK2, and TYK2) and 2 STAT members (STAT3 and STAT5). The temporal kinetics of STAT3 and STAT5 activation are markedly distinct; whereas STAT3 stays activated on prolonged G-CSFR activation for hours (or even days), activation of STAT5 peaks after 15 minutes and then declines rapidly. The gene encoding the suppressor of cytokine signaling 3 (SOCS3) is the most prominent direct transcriptional target of STAT3 in myeloid progenitors ; G-CSF–induced STAT5 targets in myeloid precursors remain ill-defined.
The cytoplasmic domain of the G-CSFR comprises 4 conserved tyrosine (Y) residues, which in the phosphorylated (p) state serve as binding sites for Src homology 2 (SH2) domain-containing signaling proteins. These pY-coupled pathways are dispensable for G-CSF–induced granulopoiesis but orchestrate the signaling output of the G-CSFR, thereby controlling the proliferation/differentiation balance in neutrophil production under basal and emergency conditions. For instance, pY729 (pY728 in mice) is a recruitment site for the inhibitory SOCS3 protein, whereas the adapter proteins Shc and Grb2, involved in proliferation and prosurvival signaling through activation of p21Ras and PI-3K/Akt, bind to pY764 (pY763 in mice). Details of pY-linked signaling pathways activated by the G-CSFR have been reviewed earlier and are summarized in Fig. 1 . Interestingly, SOCS3 does not only act in a classical negative feedback loop to attenuate G-CSF signaling, but is also involved in a transmodulatory role of the proinflammatory cytokine interferon-gamma (IFNγ) in the control of myelopoiesis. While enhancing monocytic differentiation through activation of the transcription factors IRF8 and PU.1 in myeloid progenitor cells, IFNγ reduces G-CSF–driven neutrophil differentiation via STAT3-mediated induction of SOCS3.
G-CSF responses and G-CSFR defects in severe congenital neutropenia
Severe congenital neutropenia (SCN) is characterized by a promyelocytic maturation arrest in the bone marrow, leading to defective neutrophil production and life-threatening opportunistic bacterial infections. The underlying genetic defects causing this heterogeneous disease have been partly elucidated. Mutations in the gene encoding neutrophil elastase ( ELANE ) are found in autosomal dominant and sporadic cases of SCN, whereas mutations in the gene encoding HCLS1-associated protein X1 ( HAX1 ) are responsible for the autosomal recessive form of SCN. HAX1 is a mitochondrial protein and a prevailing hypothesis is that increased apoptosis of myeloid progenitor cells is the major underlying cellular principle responsible for the paucity of mature neutrophils in HAX1-SCN. How ELANE mutations fit into this hypothesis is not obvious. A number of possible explanations as to how mutant neutrophil elastase (NE) proteins cause neutropenia have been put forward. A current view is that mutant NE causes an unfolded protein response (UPR) in the endoplasmic reticulum (ER), leading to cell damage and apoptosis. This hypothesis is supported by a recent study showing that mutant NE triggers the UPR in a transgenic mouse model and that inhibition of ERAD (ER-associated degradation) by the proteasome inhibitor bortezomib, allowing misfolded NE proteins to accumulate in cells, gave rise to reduced neutrophil levels. Whether these experimental conditions bear significance for clinical SCN remains to be determined. G-CSF is used successfully in the clinic to alleviate neutropenia in patients with SCN, but there is a significant variation in dosage requirements and efficacy. Questions relevant for this review are how G-CSF therapy reverts the neutropenic phenotype and why G-CSF responses among patients are variable, even within the same genetic subtype of SCN.
A major clinical complication in SCN is the progression toward myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), emerging with the increased life expectancy owing to the reduced infection-related mortality. In the 1990s, the first reports appeared in which nonsense mutations in the gene encoding the G-CSFR were described that result in the truncation of approximately 100 amino acids of the cytoplasmic domain of the receptor protein and that were associated with leukemic progression of the disease. Studies in cell line and knock-in mouse models revealed that these truncated G-CSFRs were hampered in their ability to transduce signals controlling neutrophil differentiation in vitro and conferred hyperproliferative responses to G-CSF in vivo. The mutations truncating the G-CSFR protein are acquired in hematopoietic clones during the neutropenic phase. These clones may expand and give rise to leukemia, a process that will be further discussed later in this review. The truncation mutants are hampered in internalization and lack Y729-containing binding motif for SOCS3 (see Fig. 1 ). Activation of the G-CSFR truncation mutants results in drastically altered kinetics of STAT5 activation, that is, from transient (minutes) to sustained (hours) activation, and increased production of intracellular reactive oxygen species (ROS). Importantly, STAT5 is essential for the clonal expansion of hematopoietic stem and progenitor cells in mice harboring the truncating G-CSFR-d715 mutation. The elevated G-CSF–induced ROS levels in bone marrow cells expressing truncated G-CSFRs may potentially contribute to abnormal signaling by several mechanisms: by inactivation of oxidation sensitive phosphatases that negatively control growth factor signaling, by inducing responses to cellular stresses caused by ROS-induced damage, and by causing DNA damage leading to an elevated mutation rate in the HSC compartment. In the next paragraphs, these possibilities and their potential consequences for leukemic evolution of SCN are further discussed.
Lysosomal routing of the G-CSFR controlled by receptor ubiquitination
After activation of the G-CSFR at the plasma membrane, the G-CSF/G-CSFR complex is internalized. A conserved dileucine-containing stretch (STQPLL) at amino acid positions 749 to 754 within the G-CSFR cytoplasmic region is the major determinant responsible for clathrin-mediated receptor endocytosis. The internalized G-CSF/G-CSFR complex then enters the early endosome (or sorting endosome) compartment, where receptor complexes can undergo 2 fates: processing for lysosomal degradation or rerouting toward the plasma membrane. Once present in early endosomes, the major portion of the G-CSF/G-CSFR complexes will be incorporated into so-called multivesicular bodies (MVBs). This process is governed by the endosomal sorting complex required for transport (ESCRT) machinery and involves ubiquitination on lysine (K) residues as a major protein modification, resulting in the interaction of the G-CSFR with multiple ubiquitin-binding adaptor proteins within the ESCRT complex. A G-CSFR mutant lacking cytoplasmic lysine residues (mutant K5R) accumulates in early endosomes, emphasizing that lysine ubiquitination of the G-CSFR is crucial for routing toward late endosomes and lysosomes. Additional mutational analysis showed that only the most membrane proximal lysine residue, K632, is critical for lysosomal routing. Ubiquitination is mediated by enzyme complexes consisting of ubiquitin-activating (E1), conjugating (E2), and ligating (E3) enzymes. The ring finger protein c-Cbl is the major E3 ligase involved in the ubiquitination of epidermal growth factor receptor (EGFR) and a number of other receptor tyrosine kinases. c-Cbl has been implicated in the ubiquitination of some receptors of the cytokine receptor superfamily. Alternative E3 ligase systems implicated in cytokine receptor ubiquitination include the Skp, Cullin, and F-box beta-Transducin repeat containing protein (beta-Trcp) and receptor-associated ubiquitin ligase (RUL) ; however, neither of these E3-ligases has been implicated in lysosomal routing of the G-CSFR and, instead, a role for SOCS3 has been proposed.
SOCS3 contains 2 distinct functional domains, the kinase inhibitory region (KIR), inhibiting JAK activity, and the SOCS box. The SOCS box recruits the E3 ligase complex consisting of Elongin B/C, Cullin 5, and the RING finger protein Rbx2. The SOCS3-SOCS box inhibits G-CSF responses in vitro and in vivo and siRNA-mediated knockdown of SOCS3 expression reduces the ubiquitination of the membrane proximal lysine residue K632 of the G-CSFR. Furthermore, like mutant K632R, the G-CSFR mutant Y729F that is unable to bind SOCS3 was hampered in lysosomal routing. Finally, the inhibitory effects of SOCS3 on G-CSF–induced STAT5 activation in reporter assays depended on the presence of G-CSFR-K632. Taken together, these results suggest that SOCS3 controls lysosomal routing of G-CSFR by a mechanism involving the membrane proximal K632, a residue that is highly conserved in evolution and that is shared by multiple cytokine receptors.
Protein ubiquitination is counteracted by de-ubiquitinating enzymes (DUBs). Expression of a family of DUBs comprising DUB1, DUB1A, DUB2, and DUB2A is controlled by cytokines. G-CSF specifically induces the expression of DUB2A in myeloid cells, leading to de-ubiquitination and accumulation of the G-CSFR in early endosomes. Based on these findings, a model has been proposed in which SOCS3 and DUB2A act in concert to control the dynamics of lysosomal routing of G-CSFR and signaling from the early endosome compartment.
The G-CSFR truncation mutants, lacking both the STQPLL internalization motif and the tyrosine-based SOCS3 binding site (Y729), are severely hampered in internalization and lysosomal routing. In addition, they are more efficiently transported to the plasma membrane in the forward (biosynthetic) pathway. Because of their prolonged half-life and residence time at the plasma membrane, it can be immediately inferred why the G-CSFR truncation mutants act dominantly over the wild-type form. It remains a puzzle, however, why the G-CSFR mutations in SCN almost invariably affect forward and retrograde traffic of the G-CSFR. Strikingly, mutations in a number of neutropenia syndromes (eg, X-linked neutropenia [XLN], Hermansky-Pudlak syndrome type 2, Griscelli syndrome type 2, and p14 [ROBLD3] deficiency) all affect proteins involved in endosomal or lysosomal sorting. Two patients with XLN treated with G-CSF have been reported who developed AML with G-CSFR mutations similar to those found in SCN. Furthermore, HAX1 was initially identified as a protein interacting with HCLS1, a hematopoietic cell–specific cortactin-like protein that modulates endosomal trafficking through its interaction with actin. Taken together, these findings merit the hypothesis that abnormalities in endosomal and lysosomal sorting processes affect G-CSFR traffic and inhibit G-CSF responses in SCN and XLN and that G-CSF therapy favors the selection of G-CSFR mutant clones that escape this inhibition.
Impact of intracellular routing on G-CSFR function
Apart from controlling the duration and amplitude of signals, intracellular trafficking of activated receptors has been shown to contribute to diversification of signaling by certain receptor systems. Hence, an important question is how G-CSF signaling is dynamically controlled by intracellular trafficking and how this is affected in case of the internalization defective G-CSFR truncation mutants.
Local Control of G-CSF Signaling by Protein Tyrosine Phosphatase 1B
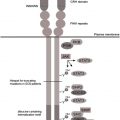
Two protein tyrosine phosphatases (PTPs) that are most frequently implicated in cytokine receptor signaling are the SH2-containing protein tyrosine phosphatase SHP1, encoded by PTPN6 , and SHP2, encoded by PTPN11 . Although SHP1 moderately inhibits G-CSFR signaling via a still unknown mechanism, a role for SHP2’s phosphatase activity has not been established. Rather, SHP2 promotes RAS/MAPK signaling by recruiting son of sevenless (SOS), a major nucleotide exchange factor for RAS, in a complex with GRB2 to the plasma membrane. The RAS promoting activity of SHP2 is enhanced by mutations in PTPN11 that cause Noonan syndrome and by PTPN11 mutations acquired in patients with myeloproliferative disease or AML. More recently, a prominent role for PTP1B in G-CSFR signaling has been discovered. PTP1B has been originally implicated in metabolism and associated diseases, that is, obesity and diabetes and receptors inhibited by PTP1B include the insulin receptor (IR), platelet-derived growth factor receptor (PDGFR), insulin-like growth factor 1 receptor, and the EGFR. In marked contrast to SHP1 and SHP2, PTP1B is not a cytoplasmic enzyme recruited to target proteins through pY-SH2 interactions, but is anchored to the outer membrane of the ER. PTP1B robustly inhibits G-CSF–induced JAK/STAT activation, phosphorylation of G-CSFR tyrosines, and proliferation of myeloid progenitors in colony assays. Confocal microscopy and in situ proximity ligation assays showed that exclusively the activated G-CSFRs in early endosomes, but not those at the plasma membrane, interacted with ER-resident PTP1B. Inhibition of G-CSF responses by PTP1B thus depends on intracellular trafficking of the activated G-CSFR ( Fig. 2 ).