Target tissue
Glucocorticoid hypersensitivity = Glucocorticoid excess
Glucocorticoid resistance = Glucocorticoid deficiency
Central nervous system
Insomnia, anxiety, depression, defective cognition
Fatigue, somnolence, malaise, defective cognition
Liver
+ Gluconeogenesis, + lipogenesis
Hypoglycemia, resistance to diabetes mellitus
Fat
Accumulation of visceral fat (metabolic syndrome)
Loss of weight, resistance to weight gain
Blood vessels
Hypertension
Hypotension
Bone
Stunted growth, osteoporosis
Inflammation/immunity
Immune suppression, anti-inflammation, vulnerability to certain infections and tumors
+ Inflammation, + autoimmunity, + allergy
Primary Generalized Glucocorticoid Resistance or Chrousos Syndrome
Primary Generalized Glucocorticoid Resistance or Chrousos syndrome is a familial or sporadic allostatic condition , which is characterized by target tissue insensitivity to glucocorticoids in almost all organs [9–16]. Because of the generalized nature of glucocorticoid resistance, all the neuroanatomic structures participating in the formation of the glucocorticoid negative feedback loops display decreased response to glucocorticoids, leading to compensatory activation of the hypothalamic–pituitary–adrenal (HPA) axis. As a result, the increased secretion of corticotropin-releasing hormone (CRH) and arginine vasopressin (AVP) from the hypothalamus into the hypophysial portal system triggers the production and release of adrenocorticotropic hormone (ACTH) by the anterior pituitary gland. Hypersecretion of ACTH results in adrenal cortex hypertrophy and triggers the production of cortisol, adrenal androgens [androstenedione, dehydroepiandrosterone (DHEA), and DHEA-sulfate (DHEAS )], and steroid precursors with mineralocorticoid activity (deoxy-corticosterone and corticosterone) [9–16].
The clinical spectrum of Chrousos syndrome is broad, ranging from completely asymptomatic cases to mild or even severe cases of mineralocorticoid and/or androgen excess . The increased concentrations of steroid precursors with mineralocorticoid activity may cause hypertension and/or hypokalemic alkalosis , while adrenal androgen excess may result in ambiguous genitalia in karyotypic females, precocious puberty, acne, hirsutism, male-pattern hair loss and hypofertility in both sexes, oligo-amenorrhea and menstrual irregularities in women, and oligospermia in men [9–16]. It is worth noting that clinical manifestations of glucocorticoid deficiency are rare and have been reported in adults with chronic fatigue [14, 17, 18], in a child with hypoglycemic generalized tonic–clonic seizures during an episode of febrile illness [19], and in a newborn with profound hypoglycemia, reported easy “fatigability” with feeding and growth hormone deficiency [20]. Interestingly, the increased concentrations of CRH may account for anxiety and depression in some patients with Chrousos syndrome [16].
The aforementioned clinical heterogeneity of Chrousos syndrome occurs because of differences in target tissues’ sensitivity to glucocorticoids, mineralocorticoids, and adrenal androgens among patients [9–16]. In addition to their cognate receptors, other molecules participating in steroid signaling pathways , such as hormone inactivating or activating enzymes, immunophilins, and heat shock proteins, as well as genetic and epigenetic factors contribute substantially to tissue response to steroid hormones [13, 15, 16].
The Molecular Basis of Chrousos Syndrome
The molecular basis of Chrousos syndrome has been ascribed to point mutations, insertions or deletions in the NR3C1 gene , which encodes the human glucocorticoid receptor (hGR) [9–16]. The NR3C1 gene is located on the short arm of chromosome 5 and contains 10 exons. Exons 2–9 express all the protein isoforms, whereas exon 1 consists of several promoters, which enable the initiation of transcription in a promoter- or tissue-specific fashion [4, 5, 16, 21]. The alternative splicing of exon 9 gives rise to the two main protein isoforms , the hGRα and the hGRβ, which have distinct properties with respect to localization, ligand-binding ability, and transcriptional activity [22–26]. Moreover, the alternative splicing of the NR3C1 gene generates three more receptor subtypes, the hGRγ, hGR-A, and hGR-P [23]. At the mRNA level, the alternative translation initiation of hGRα generates eight receptor isoforms α (hGRα-A, hGRα-B, hGRα-C1, hGRα-C2, hGRα-C3, hGRα-D1, hGRα-D2, and hGRα-D3) and possibly eight β isoforms as well, with distinct intracellular localization and transcriptional activity [27, 28].
The classic hGRα belongs to the steroid hormone receptor family of the nuclear receptor superfamily and functions as a ligand-induced transcription factor influencing the transcription rate of numerous genes [4, 5, 16]. At the protein level, the hGRα consists of four functional domains : (1) the N-terminal or immunogenic (NTD) , which contains important amino acids that undergo several posttranslational modifications; (2) the DNA-binding domain (DBD), which consists of the characteristic and highly conserved motif of two zinc fingers, and enables the interaction between the receptor and its target DNA sequences in the glucocorticoid-responsive genes; (3) the hinge region, which provides the appropriate structural flexibility to the protein and allows the receptor to interact with different target genes; and (4) the ligand-binding domain (LBD) , which is responsible for the binding of the receptor to glucocorticoids and contains sequences important for the translocation of the protein from the cytoplasm to the nucleus following activation, as well as amino acids that mediate the interaction of the receptor with coactivators in a ligand-dependent fashion [4, 5, 16].
At the target cell, the glucocorticoid signaling pathway is activated upon the binding of the receptor to synthetic and/or natural glucocorticoids, which causes the appropriate conformational changes to the protein, enabling the receptor to dissociate from chaperon heat shock proteins (HSPs) and immunophilins, and to translocate into the nucleus [4, 5, 16]. Within the nucleus, the activated receptor forms homo- or heterodimers and binds to the specific glucocorticoid response elements (GREs) within the promoter sequences of target genes, thereby inducing or repressing the transcription of the latter. Furthermore, the ligand-bound hGRα can modulate gene expression independently of DNA binding by physically interacting with other fundamental transcription factors, such as the activator protein-1 (AP-1), nuclear factor-kB (NF-kB), and signal transducers and activators of transcription (STATs) [4, 5, 16].
Patients with Chrousos syndrome usually harbor a point mutation, insertion or deletion in the NR3C1 gene, which generally results in a defective glucocorticoid receptor and impaired glucocorticoid signal transduction, leading to reduced tissue sensitivity to glucocorticoids. The majority of the reported mutations are located in the LBD (Fig. 1), leading to a broad spectrum of clinical manifestations [17, 19, 20, 29–44]. The first identified NR3C1 gene mutation was an adenine to thymine substitution at nucleotide position 1922, which resulted in substitution of aspartic acid to valine at amino acid residue 641 at the LBD [17]. Within the last four decades, the tremendous progress of molecular and structural biology has provided us with the appropriate methods to study in depth the molecular mechanisms of action of the mutant glucocorticoid receptors.
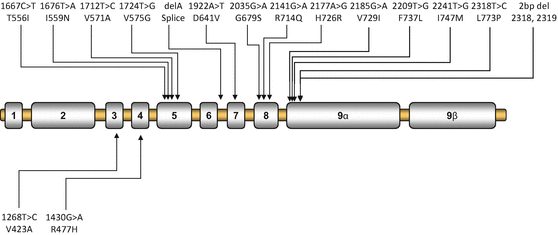

Fig. 1
Schematic representation of the known mutations of the NR3C1 gene causing Chrousos syndrome. Mutations in the upper panel are located in the LBD of the receptor, while the V423A and R477H mutations are located in the DBD of the receptor
From the Bedside to the Bench: Molecular and Structural Biology of Chrousos Syndrome
We and others have thoroughly investigated the molecular mechanisms of action of the defective natural hGRs [17, 19, 20, 29–44]. We systematically investigated: (1) the transcriptional activity of the mutant receptors through reporter assays; (2) the protein expression via Western blotting; (3) the ability of the mutant receptors to exert a dominant negative effect upon the hGRα-mediated transcriptional activity using reporter assays; (4) the ability of the mutant receptors to transrepress the NF-kB signaling pathway using reporter assays; (5) the affinity of the mutant receptors for the ligand through dexamethasone-binding assays; (6) the subcellular localization of the mutant receptors and the time required to translocate from the cytoplasm to nucleus following exposure to the ligand using green fluorescent protein (GFP)-fused plasmids; (7) the ability of the mutant receptors to bind to GREs via in vitro binding assays; (8) the interaction of the mutant receptors with the glucocorticoid receptor-interacting protein 1 (GRIP1) coactivator using Glutathione-S-Transferase (GST) pull-down assays; and (9) the conformational change of the mutant receptor that causes glucocorticoid resistance by structural biology studies. The molecular defects of the mutant receptors that have been identified in patients with Chrousos syndrome are presented in Table 2 [17, 19, 20, 29–44].
We have recently identified a novel point mutation in the NR3C1 gene associated with Chrousos syndrome in a patient that presented with hirsutism, acne, alopecia, anxiety, fatigue, and irregular menstrual cycles, but no clinical manifestations suggestive of Cushing’s syndrome [44]. The patient harbored a novel A>G transition at nucleotide position 2177, which resulted in histidine (H) to arginine (R) substitution at amino acid position 726 of the receptor [44]. Following identification, we applied the abovementioned methods in an attempt to investigate how the mutant receptor hGRαH726R caused glucocorticoid resistance. Compared with the wild-type receptor, the hGRαH726R displayed reduced ability to transactivate target genes and to transrepress the NF-kB signaling pathway, had 55 % lower affinity for the ligand and a fourfold delay in cytoplasmic-to-nuclear translocation, and interacted with the GRIP1 coactivator mostly through its activation function-1 domain [44] (Fig. 2). Finally, structural biology studies showed that the H726R mutation revealed a significant structural shift in the rigidity of helix 10 of the receptor, which caused reduced flexibility and decreased affinity of the mutant receptor for the ligand [44] (Table 2).
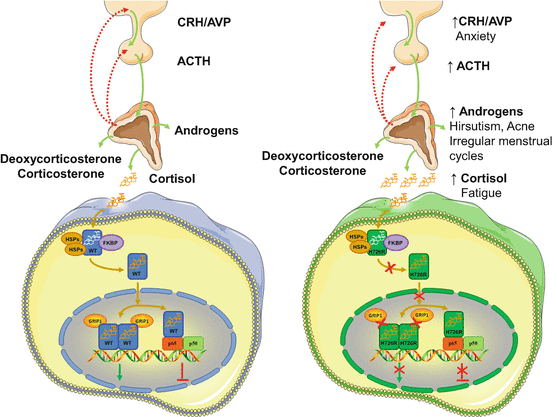

Fig. 2
Molecular mechanisms of action of the mutant receptor hGRαH726R causing Chrousos syndrome. Both the wild-type hGRα and the mutant receptor hGRαH726R reside in the cytoplasm in the absence of ligand by forming a heterocomplex with heat shock proteins (HSPs) and FKBP51 (FKBP). Upon ligand binding, the wild-type hGRα dissociates from the heterocomplex and translocates into the nucleus, while this process of the mutant hGRαH726R is significantly delayed due to decreased ligand binding and/or impaired nuclear translocation. The wild-type hGRα induces or represses the transcriptional activity of glucocorticoid target genes by attracting to GREs several coactivators including the glucocorticoid receptor-interacting protein 1 (GRIP1), or by interacting with other transcription factors, such as the NF-kB. On the other hand, the mutant receptor hGRαH726R has impaired interaction with the GRIP1, and displays reduced ability to transactivate glucocorticoid-responsive genes and to transrepress the NF-kB signaling pathway. FKBP: immunophilins; GRIP1: glucocorticoid receptor-interacting protein 1; H726R: human glucocorticoid receptor H726R; HSP: heat shock proteins; p65: transcription factor p65; p50: transcription factor p50; WT: wild-type human glucocorticoid receptor
Table 2
Mutations of the human glucocorticoid receptor gene causing Chrousos syndrome
Author (Reference) | Mutation position | Molecular mechanisms | Genotype | Phenotype | |
|---|---|---|---|---|---|
cDNA | Amino acid | ||||
Chrousos et al. [17] | 1922 (A → T) | 641 (D → V) | Transactivation ↓ | Homozygous | Hypertension |
Hurley et al. [30] | Affinity for ligand ↓ (×3) | Hypokalemic alkalosis | |||
Charmandari et al. [37] | Nuclear translocation: 22 min | ||||
Abnormal interaction with GRIP1 | |||||
Karl et al. [31] | 4 bp deletion in exon-intron 6 | hGRα number: 50 % of control | Heterozygous | Hirsutism | |
Male-pattern hair loss | |||||
Inactivation of the affected allele | |||||
Menstrual irregularities | |||||
Malchoff et al. [32] | 2185 (G → A) | 729 (V → I) | Transactivation ↓ | Homozygous | Precocious puberty |
Charmandari et al. [37] | Affinity for ligand ↓ (×2) | Hyperandrogenism | |||
Nuclear translocation: 120 min | |||||
Abnormal interaction with GRIP1 | |||||
Karl et al. [29] | 1676 (T → A) | 559 (I → N) | Transactivation ↓ | Heterozygous | Hypertension |
Kino et al. [33] | Decrease in hGR binding sites | Oligospermia | |||
Charmandari et al. [37] | Transdominance (+) | Infertility | |||
Nuclear translocation: 180 | |||||
Abnormal interaction with GRIP1 | |||||
Ruiz et al. [34] | 1430 (G → A) | 477 (R → H) | Transactivation ↓ | Heterozygous | Hirsutism |
Charmandari et al. [39] | No DNA binding | Fatigue | |||
Nuclear translocation: 20 min | Hypertension | ||||
Ruiz et al. [34] | 2035 (G → A) | 679 (G → S) | Transactivation ↓ | Heterozygous | Hirsutism |
Charmandari et al. [39] | Affinity for ligand ↓ (×2) | Fatigue | |||
Nuclear translocation: 30 min | Hypertension | ||||
Abnormal interaction with GRIP1 | |||||
Mendonca et al. [35] | 1712 (T → C) | 571 (V → A) | Transactivation ↓ | Homozygous | Ambiguous genitalia |
Charmandari et al. [37] | Affinity for ligand ↓ (×6)
Stay updated, free articles. Join our Telegram channel
Full access? Get Clinical Tree
 Get Clinical Tree app for offline access
Get Clinical Tree app for offline access

| ||||