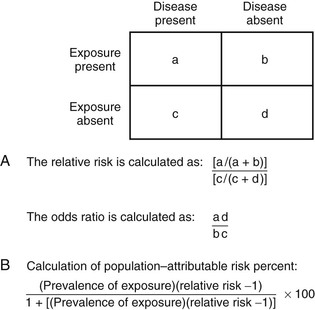
Michael T. Osterholm, Craig W. Hedberg Epidemiology is the study of health-related events in defined human or animal populations. These events include specific diseases and conditions as well as the exposures and host factors that contribute to their occurrence. The science of epidemiology was originally derived from the study of epidemics and has now been broadened to encompass all phenomena related to health in populations.1 Simply stated, epidemiology involves the careful description of events within populations and the comparison of rates at which these events occur between groups within those populations. Similar concepts and methods of epidemiology apply to both infectious and noninfectious diseases.2 The strength and adaptability of epidemiologic methods come from their underlying simplicity. For example, John Snow’s application of epidemiologic study methods led to the classic intervention of pulling the handle from the Broad Street pump during an outbreak of cholera in London in 1851. His work was based on a careful description of his observations and on a quantitative approach in analyzing the occurrence of cholera among the citizens of London. The influence of his work led to legislation mandating that all water companies in London filter their water. Of note, it was not until 1883 that Robert Koch discovered Vibrio cholerae.3 As applied to infectious diseases, at least 10 goals of epidemiologic analysis can be listed: 1. Describe patterns of infection and disease occurrence in populations. 2. Identify outbreaks or unusual rates of disease occurrence. 3. Facilitate laboratory-based efforts to identify infectious agents. 6. Assist in the understanding of disease pathogenesis. 8. Develop and evaluate treatment protocols through clinical trials. 10. Describe and assess the use of prevention measures on a community-wide basis. These comprehensive goals far exceed the often-considered goal of epidemiologic analysis to investigate and control epidemics or outbreaks. The goals of epidemiologic analysis can be illustrated by a historical review of the unfolding of the human immunodeficiency virus (HIV) epidemic. After the acquired immunodeficiency syndrome (AIDS) was initially described in 1981, a national epidemiologic surveillance case definition was developed. Disease surveillance was initiated to characterize the cases by standard measures of time, place, and person and to identify population groups at risk. Based on these efforts, an infectious etiology was hypothesized early in the epidemic, before the first laboratory evidence of an etiologic agent was presented. Combined clinical, epidemiologic, and laboratory studies led to the identification of HIV as the cause of AIDS and to the development of sensitive and specific serologic tests for infection. This progress in turn led to studies that characterized the spectrum of illness associated with HIV infection. Epidemiologic studies of persons infected with HIV (with or without AIDS) have characterized the routes of HIV transmission, have shown that the occurrence of other sexually transmitted infections can increase the risk of HIV transmission, and have demonstrated that HIV infection can enhance the transmission of other agents, such as Mycobacterium tuberculosis. Longitudinal follow-up studies of HIV-infected persons have identified long-term survivors—individuals who have been infected for more than 10 years (and now more than 30 years) and have received no treatment yet remain without disease.4 Others have been studied who were exposed to HIV on numerous occasions but did not become infected. Collectively, these studies provided important observations leading to better understanding of the mechanisms of resistance to HIV infection and disease. Clinical trials were conducted to assess the efficacy of antiretroviral agents and combinations of drugs to increase the effectiveness of therapy and reduce the rate of resistance to individual drugs. Development of potential HIV vaccines progressed to the implementation and innovative design of phase III human trials.5,6 Other trials were conducted to assess the efficacy of a range of antimicrobial agents aimed at preventing a variety of opportunistic infections. Finally, community-based programs were developed on the basis of epidemiologic data to promote behavior change aimed at reducing the risk of HIV transmission. Epidemiologic methods were also applied to evaluate these community-based programs and to establish a framework in which a disease-modifying HIV vaccine could be integrated into a comprehensive HIV prevention program. These examples illustrate the broad range of roles that epidemiologic methods have played in understanding and controlling the HIV epidemic. During 2003, the global application of combined clinical, epidemiologic, and laboratory studies led to the rapid detection, characterization, and, ultimately, control of an epidemic of severe acute respiratory syndrome (SARS) caused by a novel coronavirus.7–9 Epidemiologic studies identified the original source of transmission from palm civets to humans through wild-animal markets in China and demonstrated the global spread of the epidemic through person-to-person transmission.10 The eradication of the epidemic strain from humans and the identification of the wildlife reservoir of SARS coronaviruses established a framework for preventing future SARS outbreaks. The global response to SARS serves as a model for the usefulness of epidemiologic methods. In 2012, the emergence of human infections caused by another novel coronavirus in the Middle East, the Middle East respiratory syndrome coronavirus (MERS-CoV) again resulted in the combined rapid conduct of clinical, epidemiologic, and laboratory studies to reduce the risk of this virus causing another SARS-like epidemic.11–14 As of August 1, 2013, there have been 94 laboratory-confirmed cases of infection with MERS-CoV, including 46 deaths. The lessons learned from similar studies conducted in 2003 have been invaluable in responding to the occurrence of MERS-CoV 2012 to 2013 (see Chapter 157). On March 31, 2013, the public health authorities of China reported three cases of laboratory-confirmed human infection with a novel avian influenza A (H7N9) virus. By late May 2013, approximately 2 months after the initial report, the number of laboratory-confirmed H7N9 infections reached 132, with 37 deaths.15 The rapid conduct of clinical, epidemiologic, and laboratory studies were initiated by Chinese medical and public health officials with support from experts from around the world. It was determined that the novel H7N9 viruses were reassortants, comprising H7 HA, N9 NA, and the six internal genes of H9N2 influenza A viruses. This combination of influenza genes had not previously been identified among viruses obtained from birds, humans, or any other species, although individual genes are related to those of recent avian influenza viruses circulating in East Asia. H7N9 viruses obtained from human cases, poultry, and environmental samples were closely related and contained a number of genetic signatures previously associated with low pathogenicity in poultry, enhanced capacity for mammalian infection, and resistance to the adamantane class of antiviral drugs.16,17 The detection of H7N9 virus in live poultry markets in the vicinity of human cases in Shanghai, the contact history with live poultry or live poultry markets in a substantial number of cases, and the major reduction in human cases after the closure of live poultry markets throughout eastern China, suggest exposure to live poultry as a key risk factor for human H7N9 infection18 (see Chapter 157). An essential aspect of any epidemiologic study is careful definition of the infection, disease, condition, or factor that is being studied. Specificity and sensitivity are concepts that are frequently used in reference to laboratory test performance, particularly with tests that are used for screening purposes.1 However, in the epidemiologic study of infectious diseases, it is important to also apply the concepts of specificity and sensitivity more broadly in terms of diagnosis of infection and disease. For example, the diagnosis of smallpox was both highly specific and sensitive. Few other diseases could be confused with smallpox (i.e., the diagnosis was specific), and clinical disease developed in most people who became infected with smallpox virus (i.e., the diagnosis was sensitive). These qualities, in addition to the facts that humans were the only important reservoir for the smallpox virus and that highly immunogenic vaccines had been developed, led to the successful eradication of smallpox.19 In contrast, many clinical conditions or syndromes, such as diarrhea, are caused by more than one etiologic agent. Epidemiologic studies of diarrheogenic Escherichia coli are complicated by the fact that diarrhea is not specific for E. coli, and the sensitivity of E. coli detection is limited due to an array of virulence factors that can result in disease yet are not detected by standard biochemical tests.20 Even the ability to detect specific agents and virulence factors by rapid, nonculture methods does not resolve these difficulties.21 E. coli O157:H7 and some other Shiga-toxin–producing E. coli (STECs) may lead to a broad spectrum of clinical illnesses, including uncomplicated diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome. However, not all STEC strains have the same disease-causing potential. Depending on whether the goals of a particular study address the clinical illness, the specific agent, or the public health implications of detecting the agent in clinical, food, or environmental samples, investigators may choose a case definition that casts a wide net or is more narrowly focused. The type of definition can have a substantial impact on study results and should be carefully considered before a specific study is undertaken. Epidemiologic studies may be designed to evaluate outcome variables other than infection or disease occurrence. In these situations, how the outcome variables and study population are defined and measured can affect interpretation of the results and the validity of the conclusions. For example, in the development of recombinant vaccines for hepatitis B virus (HBV), two vaccine formulations, containing either 10 µg or 20 µg of hepatitis B surface antigen (HBsAg) in each dose, were evaluated in clinical trials. Higher antibody titers developed in subjects who were administered vaccine with the higher dose. Both vaccines produced sufficient levels of antibody to be considered protective against infection, and both were licensed by the U.S. Food and Drug Administration. However, when the vaccines were more broadly administered to Minnesota hospital employees, those who received vaccine with 20 µg HBsAg per dose were more likely than those given the lower-dose vaccine to have detectable antibody when tested within 6 months after completing the three-dose series.22 The results of this investigation suggested that sociodemographic factors of the community-vaccinated population, such as age, gender, weight, and smoking, affected the outcome of vaccination programs in ways that were not predicted by the clinical trials.23 In a study of children with inflammatory bowel disease, only 56% of children previously vaccinated against HBV had immunity to HBV, a mean of 13 years later.24 Older age, lower albumin levels, and the presence of pancolitis were associated with lack of immunity. The use of immunosuppressive therapy was not associated with a lack of immunity, but it was associated with a lack of response to an additional dose of HBV vaccine given to children who lacked immunity.24 Establishing specific enrollment criteria for cases of infection or disease in epidemiologic studies is critical to obtaining valid and biologically meaningful results. For example, large multistate outbreaks of E. coli O157:H7 have been documented with increasing frequency. However, without molecular subtyping of E. coli O157:H7 strains, population-based surveillance is limited in its ability to detect and determine when an unexpected number or temporal clustering of cases actually documents a common vehicle-associated outbreak. In September 2006, epidemiologists and public health laboratory workers in Wisconsin noticed a small cluster of E. coli O157:H7 cases with a common pulsed-field gel electrophoresis (PFGE) subtype pattern. The Wisconsin state laboratory posted the PFGE pattern on the national subtyping network for foodborne disease surveillance known as PulseNet. Within 1 week, multiple states reported matching cases, an epidemiologic study was conducted to evaluate case exposures, and fresh spinach was identified as the vehicle.25 Ultimately, 205 cases in 26 states were linked to the outbreak, and the results of the investigation stimulated important investments in research related to the safety of leafy-green produce items. Because of its discriminatory ability, the Centers for Disease Control and Prevention (CDC) has adopted PFGE as the standard molecular subtyping method for national surveillance.26 CDC-PulseNet now commonly plays an important role in identifying and investigating multistate outbreaks of foodborne disease. A similar issue regarding the definition of cases and the population in which they occur confronts public health officials when they must consider intervention activities because of a possible outbreak of certain infectious diseases. It is common practice to define outbreaks as the occurrence of cases of disease at a frequency greater than expected.1 When an outbreak occurs, it is necessary to define the population at risk (i.e., the denominator) if an accurate measure of the rate of disease is to be calculated. For example, it is not unusual to recognize a cluster of cases of Neisseria meningitidis disease in the community in populations not previously vaccinated. Because outbreaks of invasive N. meningitidis disease are known to occur in closed populations, such as persons living in dormitories and barracks, and because a vaccine and antibiotic chemoprophylaxis are available to prevent or control these outbreaks, the occurrence of multiple cases of meningococcal disease inevitably prompts a rapid public health assessment.27 Cases of meningococcal disease tend to occur during well-described seasonal peak periods, so it is possible that a cluster of unrelated cases may occur in a defined population. Conversely, a common strain may be transmitted within social networks that form a population group that is not easy to define. During 2005 to 2006, 23 cases of serogroup C meningococcal disease occurred among illicit drug users and their contacts in Brooklyn, New York.28 From 2010 to 2012, 18 men who have sex with men (MSM) in New York City were infected by a closely related strain.29 The need for public health intervention is quite different for a cluster of cases representing an outbreak associated with a single strain, compared with a cluster in which each case is caused by a different group or strain of N. meningitidis.30 However, in many situations, strains are not available for further subtyping, because laboratory capacity to distinguish strains is limited. As illustrated above, a companion problem to the definition of cases is definition of the population at risk. To determine whether cases of disease are occurring at a frequency greater than expected, it is necessary to consider baseline incidence rates of disease. During the outbreak of meningococcal disease among illicit drug users, the population-based rate of illness in Brooklyn never approached the threshold for public health intervention of 10 cases per 100,000 population over a 3-month time period.27,28 Although it was not possible to enumerate the actual population at risk, the ongoing occurrence of cases in persons with similar histories led to empirical judgments that an outbreak was occurring and a vaccination program targeting illicit drug users was needed.28 In the outbreak involving MSM, data from a community health survey was available to estimate the population of MSM and determine that the risk of meningococcal disease among MSM was 80 times the risk among non-MSM.29 Timely decisions regarding major community-based interventions after the observation of a cluster of meningococcal diseases often are made without adequate information regarding the status of a possible outbreak. Similar situations occur with other pathogens as well. Two common measures of the occurrence of disease in populations are incidence and prevalence.1 Incidence represents the occurrence of new cases of infection or disease per unit of population per time period. It is common to express incidence rates in terms of person-years of exposure. Prevalence describes the number of current cases of disease per population unit at the time of observation. The relationship between incidence and prevalence depends on the duration of infection or disease. For example, the incidence of Lyme disease or hepatitis A virus (HAV) infections over a period of 1 year is always greater than its prevalence at a given point because the disease has a very short duration. In contrast, the prevalence of HIV or M. tuberculosis infections is always greater than its incidence because the infection is chronic, and infected persons may live for years after the initial infection. The results of epidemiologic studies to compare the risk of infection or disease and the presence or absence of specific risk factors are presented in terms of relative risk and odds ratios. Relative risk (RR) is the ratio of the rate of illness or infection among persons who were exposed to the rate among persons who were not exposed (Fig. 13-1). RRs may also be called rate ratios and are the products of cohort studies. In case-control studies, odds ratios (ORs) are determined and approximate the relative risk. ORs provide a valid estimate of the RR under conditions that prevail in most case-control studies: the cases of disease are newly diagnosed, prevalent cases are not included in the control group, and the selection of cases and controls is not based on exposure status.31 An increased RR or OR (i.e., >1.0) for an exposure variable indicates that the exposure is related to an increased risk of disease. Similarly, a decreased RR or OR (i.e., <1.0) indicates that the exposure variable is related to a decreased risk of disease. For example, the consumption of undercooked ground beef has been associated with an increased risk of E. coli O157:H7 infection in outbreak settings and of sporadic E. coli O157:H7 infections in the community.32 Although RRs and ORs do provide a measure of the risk of disease associated with a specific factor, they do not directly describe how much disease in the community can be attributed to that factor. Rather, the attributable risk or fraction considers both the RR for an exposure variable and the proportion of the population exposed to that variable. In a case-control study of sporadic E. coli O157:H7 infections conducted by the Foodborne Disease Active Surveillance Network (FoodNet) in 1996 to 1997, persons who ate undercooked hamburgers away from home had approximately a 6 times greater risk of E. coli O157:H7 than those who did not. For those who ate undercooked hamburger at home, the risk was only 2 times greater. However, eating undercooked hamburger at home was more common than eating it away from home. Therefore, eating undercooked hamburger at home accounted for an estimated 8% of cases, whereas the riskier (i.e., higher OR) practice of eating undercooked hamburger away from home accounted for 7% of cases. Furthermore, persons who ate at a table service restaurant were only 1.7 times as likely to have E. coli O157:H7 infection than those who did not. But, because eating at a table service restaurant is a very common practice and therefore represents more frequent exposure, it accounted for an estimated 20% of cases. Although it had the weakest statistical association, it accounted for the highest proportion of cases. Similarly, in a case-control study of sporadic listeriosis cases reported in FoodNet sites from 2000 to 2003, living on a cattle farm had the strongest association with illness (OR = 13.75), but, because it was an uncommon exposure, it had a population attributable fraction of only 1.6%. However, eating melons at a commercial establishment accounted for 10.6% of cases, even though the OR was only 2.6.33 The identification of melon as a risk factor for sporadic listeriosis in this study subsequently made it easier for investigators to identify cantaloupe as the source for a large multistate outbreak of listeriosis in 2011.34 As these examples show, both RR and attributable risk are important measures for describing the epidemiology of infectious diseases and determining public health priorities. In the epidemiology of infectious diseases, many factors are evaluated to determine their relationship or association with a specific disease. Statistical associations, both positive and negative, may represent a true causal relationship, a confounding relationship with another factor, or a chance occurrence. If more than one factor is statistically associated with infection or disease status in univariate (single-variable) analyses, the relationship between individual factors and infection or disease status can be evaluated by multivariate regression analysis.35 These procedures allow the investigator to simultaneously control for a combination of factors in the analysis and to determine whether any of the risk factors are associated with infection or disease status independently of other factors. Another critical way of distinguishing causation from confounding or chance is by assessing the biologic plausibility of the association. An unexpected statistical association found in conjunction with an epidemiologic study may result in new understanding of how agent transmission or disease occurs. The temptation to stretch the plausibility of biology to provide meaning to statistical results is a constant danger. However, such results may be a useful guide to evaluate new hypotheses in future studies. Furthermore, “statistically significant results” may be unimportant from a disease control or a practical perspective. Statistical significance, which has historically been considered to be an event that would happen less than 1 in every 20 instances by chance alone (i.e., P < .05), represents a combination of the sample size and the strength or degree of the association. Studies with a large number of persons enrolled can produce statistically significant results for weak associations (i.e., RRs or ORs greater than 1 but less than 2), whereas studies with a limited number of enrollees may not be able to produce statistically significant results even for moderately strong or increased associations (i.e., RRs or ORs > 5). The clinical trial is cited as the gold standard of epidemiologic research. However, many epidemiologic studies cannot take place under such rigorously controlled conditions. Taking advantage of opportunities to study diseases in clinical and community settings is one of the strengths of epidemiology. In the setting of a clinical practice, epidemiology may involve studying a series of patients, participating in multicenter trials, or being a reporting source for cases of disease to public health officials. This last aspect of epidemiologic study may be a legal obligation, but it should also be viewed as an opportunity for all practicing clinicians to participate in the practice of community-based epidemiology. Academic-based research centers are often settings for clinical trials, studies requiring newly developed laboratory methods, or studies derived from referrals to clinical specialty groups. Public health departments typically do not have direct access to or contact with patients for clinical trials, but they are responsible for surveillance of reportable diseases and the investigation of outbreaks. There has been a debate about how to distinguish public health surveillance from research and the ethical considerations of that distinction.36 Each of the described settings provides opportunities for epidemiologic studies that can make major contributions to the understanding, prevention, and control of infectious diseases. Several major constraints are confronted in the design of epidemiologic studies of infectious diseases. Time is frequently a problem in the investigation of outbreaks. The need to quickly design and conduct outbreak investigations has increased with the frequency of widespread foodborne outbreaks and concerns over the potential for intentional contamination of the food supply. This necessarily limits the investigator’s ability to fully explore the outbreak setting and can result in the loss of information. In any study involving the retrospective collection of data, information may be lost because of difficulty in recalling exposure or in verifying information about the exposure. For many infectious diseases, it may be difficult to identify sufficient numbers of cases in clinical settings to conduct meaningful epidemiologic studies. In such situations, multisite collaborative projects are often needed. For example, a CDC working group on prevention of invasive group A streptococcal (GAS) disease among household contacts concluded in 1995 that the data available from a single study conducted in Ontario, Canada, were inadequate to recommend chemoprophylaxis to household contacts.37,38 Although the Canadian study suggested an increased risk of invasive disease among household contacts, this assessment was based on only four subsequent cases in households. Based on the recommendations of the work group, a multisite study coordinated by the CDC was initiated in multiple states or areas with active surveillance of invasive GAS disease. Additional surveillance studies supported the conclusion that outbreaks of invasive disease among household contacts are rare, so that recommendations for chemoprophylaxis need to be made on an individual basis.39,40 Several schemes can be used to classify or define types of epidemiologic studies (Table 13-1). Studies can be classified as descriptive or analytic and as observational or experimental. A descriptive study is designed to describe only the existing distribution of case characteristics, without regard to causal or other hypotheses.41 For example, the results of community-based surveillance for Campylobacter infection may include a summary of all cases reported in a given year by date of onset, county of residence, age, gender, and race. An analytic study is one designed to examine associations, particularly hypothesized causal relationships. For example, a case-control study could be designed to examine whether consumption of ready-to-eat meat and poultry products is a risk factor for cases of invasive listeriosis infections identified through surveillance activities. In addition to case-control studies, cohort studies, clinical trials, and cross-sectional surveys are common types of analytic studies. In practice, most epidemiologic studies involve both descriptive and analytic elements. For example, surveillance for methicillin-resistant Staphylococcus aureus (MRSA) infections in nine surveillance sites in the United States permitted both the characterization and differentiation of community- and health care–associated infections.42 Results showed that MRSA affects certain populations disproportionately. They also demonstrated that the problem is primarily related to health care but no longer is confined to intensive care units, acute care hospitals, or any health care institution. Finally, this surveillance effort demonstrated a 26% decrease in MRSA infections in the nine surveillance areas between 2007 to 2008 and 2011. TABLE 13-1 Classification Schemes for Epidemiologic Studies A more relevant distinction can be made between observational and experimental studies. Observational studies are conducted in natural settings where changes in one characteristic are studied in relation to others without the intervention of the investigator.43 Observational studies represent the bulk of epidemiologic research because they focus on events, exposures, and diseases occurring in the population during the course of routine living conditions. In contrast, experimental studies are ones in which the study conditions are under the direct control of the investigator.43 Such studies may include randomization of subjects to treatment or placebo groups and blinding of subjects and investigators to placement status. Clinical trials are the prototypical experimental study. On a broader scale, community intervention trials can also be conducted. Disease surveillance is an ongoing process that involves the systematic collection, analysis, interpretation, and dissemination of information regarding the occurrence of diseases in defined populations for public health action to reduce morbidity and mortality.44 Surveillance can be conducted in the community and in institutional settings, where it may form the basis for an infection-prevention program. For most infectious diseases, community-based surveillance is the domain of public health departments at the local or state level. All jurisdictions require licensed physicians to report the occurrence of selected diseases to the health department.45 Typically, such diseases include sexually transmitted infections, vaccine-preventable diseases, bloodborne pathogens, tuberculosis, certain invasive bacterial diseases, and enteric infections caused by Salmonella, Shigella, E. coli O175:H7, and Campylobacter. In addition to categorical reporting, most states require reporting of disease outbreaks, regardless of the cause, and have some provision to solicit reports of new and emerging diseases. Increasingly, syndromic surveillance systems are being developed to take advantage of large data streams through electronic medical records and social media,46,47 as opposed to surveillance based on isolation of a specific infectious agent. This type of surveillance has been very useful to supplement surveillance of influenza-like illness in sentinel physician practices, nursing homes, and schools to monitor influenza activity each influenza season. Surveillance for unexplained deaths from possible infectious causes, with characterization of such deaths based on the clinical syndrome at the initial evaluation, is a way to monitor the emergence of potential new infectious disease threats.48 Finally, syndromic surveillance has been established in several large cities to serve as an early warning system for the detection of bioterrorist events.49 Although useful for tracking community-wide spread of influenza-like illness, syndromic surveillance systems have shown very little sensitivity to the occurrence of infectious disease outbreaks in these cities.50,51 Surveillance for certain pathogens has evolved to include surveillance for antimicrobial resistance. The worldwide emergence of extensively drug-resistant tuberculosis has made surveillance for drug-resistant tuberculosis routine in all jurisdictions.52 Successive waves of emergence and clonal dispersion of multidrug-resistant Salmonella Typhimurium DT 104 and multidrug-resistant Salmonella Newport among food animals and humans in the United States was detected through national surveillance to monitor resistant enteric infections.53,54 The establishment of surveillance for drug-resistant pneumococcal infections was an important step in the evaluation of the impact of the introduction of pneumococcal conjugate vaccines.55 Rates of invasive bacterial infections caused by resistant strains dropped by more than 50% after the introduction of these vaccines. The importance of surveillance for drug-resistant infections will continue to grow in the 21st century, and data collected through public health surveillance can be extremely useful to clinical care providers. Case reports for use in surveillance can be collected in an active or a passive manner. Active surveillance involves a regular, systematic effort to contact reporting sources or to review records within an institution to ascertain information on the occurrence of newly diagnosed diseases or infections. An example of an active surveillance system for foodborne illnesses is FoodNet, which operates as part of CDC’s Emerging Infections Program.56 Active laboratory-based surveillance for confirmed cases of Campylobacter, Cryptosporidium, Cyclospora, E. coli O157:H7, Listeria, Salmonella, Shigella, and Vibrio increased from five sites, covering 5% of the U.S. population in 1996 to 10 sites covering approximately 15% of the population in 2007. Each clinical laboratory in the surveillance catchment areas is contacted weekly or monthly to ensure that all confirmed infections under surveillance have been reported. These data have been extremely useful in establishing national estimates for the burden of foodborne illness in the United States and for monitoring trends in the incidence of specific foodborne agents. Passive surveillance relies on the individual clinician or laboratory to initiate the report. For many diseases of public health importance, passive surveillance can be almost as comprehensive as active surveillance. Although surveillance systems are labeled as active or passive based on how cases are reported, all surveillance systems require an active review and analysis of reported cases, with dissemination of results to key stakeholders.44 Two key qualities of community-based surveillance for infectious diseases that must be considered when interpreting surveillance data are representativeness and timeliness. These qualities vary by disease and depend on multiple factors. The first factor of importance is that the patient must seek medical attention. It is not common for persons with mild or limited illnesses to seek medical attention. Second, the physician must seek laboratory testing of appropriate clinical specimens to confirm the diagnosis. Third, the laboratory must have the capability to identify the agent. Fourth, the physician and laboratory must report the clinical and laboratory findings to public health officials in a timely manner. Fifth, the availability of molecular subtyping techniques such as PFGE and the ability to compare PFGE patterns electronically through the national computer network PulseNet can greatly increase both the sensitivity and the specificity of pathogen-specific surveillance. Even in states where laboratory-based infectious disease reporting is required, there may be confusion among physicians and laboratory officials regarding who has the responsibility for reporting. Finally, public health agencies must have the resources to conduct timely and routine follow-up of such reports, to ascertain basic case demographic and other relevant data. Failure at any step of this process results in loss of information to the community-based surveillance system. The efficiency of community-based surveillance systems varies greatly, depending on the disease, how the diagnosis is made, and the resources targeted toward the surveillance effort.44 Many emerging diseases require a diagnosis based on clinical findings, either because an etiologic agent has not been identified or because reliable diagnostic tests have not been developed. For example, for many years, the diagnosis of Lyme disease presented difficulties because many patients were not seen when the typical clinical manifestations of the disease were present, and laboratory testing was not adequate to establish the diagnosis. In contrast, the diagnosis of measles can be confirmed by specific serologic testing, regardless of whether the physician sees the patient or has the training and experience to recognize the pathognomonic clinical features of the disease. Surveillance for invasive bacterial diseases such as those caused by N. meningitidis is facilitated by the need for medical treatment because of the relative severity of the disease and the laboratory-supported diagnosis. For diseases such as these, active case ascertainment can greatly enhance the effectiveness of surveillance activities. However, active surveillance requires the commitment of personnel and other resources that are limited for many reportable diseases. Nonetheless, it may be critical to evaluating the impact of vaccines for invasive diseases such as Haemophilus influenzae type b, which declined by 95% within 6 years after the introduction of the conjugate vaccine in 1989. Active surveillance for invasive H. influenzae disease has confirmed the effective control of H. influenzae type b with no evidence of increased disease caused by non-B serotypes in young children in the United States.57 Typically, active surveillance may be conducted for a limited period when complete data are most critical. Examples include the characterization of emerging diseases such as AIDS or SARS and special surveillance projects aimed at assessing an intervention, such as evaluating whether the occurrence of intussusception was causally related to the use of rotavirus vaccine.58 Most infectious disease surveillance conducted by public health departments in the United States is passive in that it relies on the physician or the laboratory to initiate the report. Passive surveillance systems are subject to selection bias because disease reports are likely to come from a nonrepresentative sample of practicing physicians who may report specific diseases because of personal interest.44 In addition, some data (i.e., age and gender versus clinical and pathologic information) may be more readily reported because of ease of ascertainment.44 Active surveillance is relatively more common in the hospital setting. For example, surveillance of nosocomial infections is an important hospital infection-prevention activity.59 This highly specialized surveillance system has the operational advantage of a defined population, routine clinical observation of the patient population, and direct access to the laboratory. Hospital-based surveillance has been a primary epidemiologic tool in the study of drug-resistant organisms. A common type of descriptive study that is conducted in clinical settings is the case series. A case series describes the clinical features of a disease and the demographic profiles and other interesting features of patients with the disease. They are typically the domain of practicing clinicians and serve as a way of communicating significant clinical observations. For example, the SARS epidemic was first recognized outside of China as an unusual series of cases of patients with atypical pneumonia.60 As the case series grew, with evidence of transmission to hospital staff, it became apparent that an unusual outbreak was occurring. More recently, a series of 33 patients hospitalized in a medical intensive care unit during an outbreak of chikungunya virus on Reunion Island demonstrated that chikungunya virus infection can cause severe neurologic disease with the involvement of other organ systems.61 In case-control studies, persons with infection or disease are compared with controls (i.e., persons without the infection or disease under study) with respect to prior exposures likely to be related to agent transmission.1 Case-control studies by nature are retrospective, because the outcome (i.e., case status) is known at the outset of the study. Case-control studies are the most widely conducted type of epidemiologic study because they are relatively cheap, powerful, and adaptable to many settings.35 For example, in a nationwide outbreak of Salmonella enteritidis infection, the results of a case-control study identified the ice cream made by a large national producer as the source of the outbreak 10 days before S. enteritidis could be isolated from samples of the implicated ice cream.62 When S. enteritidis was isolated from the ice cream, it was shown to be present at levels of less than one to six organisms per half-cup serving, levels that rendered microbiologic surveillance of ice cream insensitive. Furthermore, the case-control study established that contamination of the pasteurized ice cream premix occurred during transport in tanker trailers that had previously carried nonpasteurized liquid eggs, even though regulatory officials were not able to isolate S. enteritidis from any environmental samples. The primary considerations in designing case-control studies are defining cases, establishing enrollment criteria, identifying suitable controls, and developing interview or other data collection processes that do not systematically result in different standards of data collection for cases versus controls. In the community setting, it is customary to select controls from the same area of residence as the cases. It is desirable for controls to resemble cases with respect to variables that are not being studied. Controls may also be matched by age, gender, or any other factor that the investigator considers necessary. For example, in studying risk factors for listeriosis, it has been important to select or match controls with a similar risk of illness based on the presence of an immunocompromising condition or treatment. This is necessary because healthy community control subjects who have exactly the same exposures are less likely to develop disease. Therefore, a case-control study of listeriosis using healthy community-based controls would require simultaneously trying to assess the risk for exposure as well as the risk for illness given exposure. However, overmatching, such as requiring the control to have the same birthday as the case, may make it difficult to identify and recruit controls. Also, once a variable is used as a matching criterion, it is no longer available for evaluation. In hospital settings, controls are frequently selected from patients with unrelated diagnoses who might otherwise be comparable to the cases. Analysis of case-control studies involves comparing exposure differences between cases and controls. Such comparison allows associations between exposure and disease to be studied even when the disease is a rare outcome of the exposure. For example, a case-control study of Guillain-Barré syndrome demonstrated an association between Campylobacter infection and Guillain-Barré syndrome.63 This association could not have been easily evaluated in a prospective cohort study because of the population size necessary to identify a similar number of cases with this syndrome. The power of the case-control methodology comes from the fact that, although illness may be an uncommon outcome of a given exposure, the common history of exposure among cases may stand in stark contrast to that among controls.
Epidemiologic Principles
Epidemiologic Study Methods
Goals of Epidemiologic Analysis
Defining Infections, Diseases, and Populations
Biology and Statistics
Determining Epidemiologic Methods Appropriate to the Study Setting
Types of Epidemiologic Studies
OBSERVATIONAL
EXPERIMENTAL
Descriptive
Surveillance
Case series
Analytic
Case-control studies
Clinical trials
Cohort studies
Community interventions
Seroincidence studies
Cross-sectional surveys
Seroprevalence surveys
Outbreak investigations
Observational Studies
Disease Surveillance
Case Series
Case-Control Studies
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Epidemiologic Principles
13