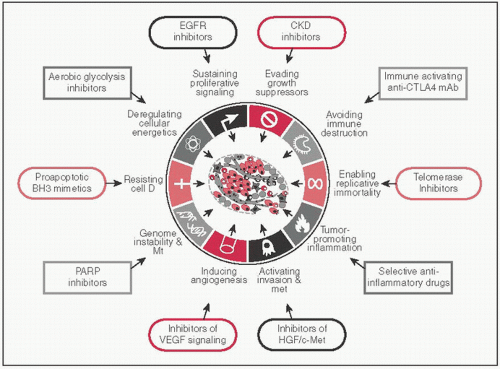
CAs share a set of common distinctive & complementary capabilities that enable tumor growth & met dissemination (Cell 2011;5:646)
Characterized by self-renewal & ability to differentiate; found in a variety of CAs including AML, CML, gliomas, breast CA (NEJM 2006;355:1253)
Two pathways involved in regulation of pattern formation during ontogeny
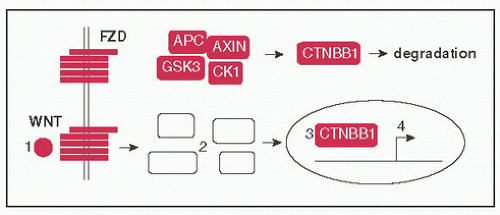
Wnt pathway: (1) WNT ligands bind to FZD (Frizzled) receptor, (2) disrupts assembly of a complex (AXIN, APC, CK1, GSK3) that tags CTNBB1 (β-catenin) for degradation, (3) CTNBB1 moves to nucleus → binds TCF/LEF freeing it from TLE (Groucho), (4) CTNBB1 & TCF/LEF activate target genes (eg, CCND1, MYC); FAP develops 2° to germline inactivating Mt of APC (Nat Rev Cancer 2013;13:11)
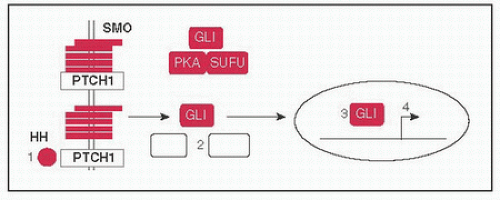
Hedgehog pathway: (1) HH ligands SHH (Sonic), DHH (Desert), & IHH (Indian) bind to PTCH1 (Patched) releasing inhibitory effect on SMO (Smoothened), (2) causes dissociation of protein complex including SUFU, PKA, & GLI, (3) GLI translocates to nucleus to (4) activate target genes; BCCs w/activating SMO Mt or inactivating PTCH1 Mt, Gorlin syndrome (BCCs, medulloblastomas, rhabdomyosarcomas) w/germline PTCH1 Mt (Nat Rev Cancer 2008;8:743)
| ||||||||||
|
Play important roles as carcinogens in several diseases (see Cancer Epidemiology Chap. for more extensive list), carcinogenesis occurs through a variety of mechanisms eg, via the actions of viral/bacterial proteins, insertional mutagenesis
Viruses: HPV (cervical CA, oropharyngeal squamous cell CAs of the head & neck, viral proteins E6 & E7 inhibit TP53 & Rb1, respectively), EBV, KSHV
Bacteria: H. pylori (gastric CA), bacterial CagA protein → activates tyrosine phosphatase SHP2 affecting various pathways involved in cell migration, adhesion, & apoptosis
Oncogenes: Contribute to the development or maintenance of the neoplastic phenotype, converted from protooncogenes w/c have nl functions that are involved in cell differentiation, proliferation, & apoptosis → these may sustain gainof-function or activating Mt that result in constitutive activation (NEJM 2008;358:502)
Oncogene addiction: Process by w/c tumor cells become highly dependent on an oncogenic protein or pathway for sustained proliferation or survival, 1° driver Mt in oncogenes are often mutually exclusive (Cancer Res 2008;68:3077)
Many oncogenes encode growth factor receptors or proteins involved in cell signaling
TSGs: Encode proteins responsible for inhibiting tumor formation or maintenance, involved in processes like DNA repair & apoptosis
Initiated by ligand binding to growth factor receptors that activate downstream pathways; mechanism by w/c an extracellular signal is transduced through a cell, resulting in a change in gene expression, modifying specific cellular processes
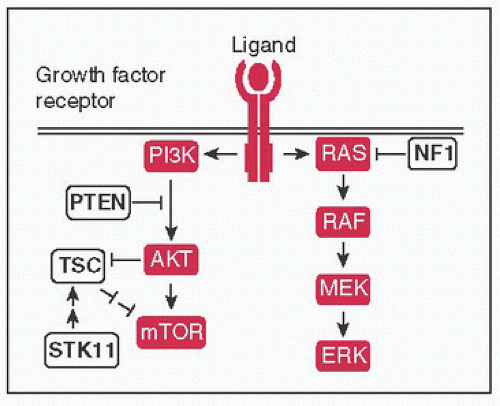
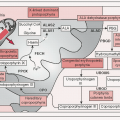
Figure 4-4 Growth factor signaling: Ligand binding (eg, EGF, TGFα, epiregulin) → receptor dimerization & phosphorylation of intracellular TK domains → downstream pathway activation; ↑ pathway activity mediated by signal transduction proteins (protooncogenes) of MAPK and PI3K pathways, negative regulators (TSGs) shown in white → ultimately affect processes such as cell survival & proliferation, differentiation, angiogenesis, etc.
Can be targeted by mAbs (eg, bevacizumab for VEGF, cetuximab for EGFR, see Monoclonal Antibodies Chap.) or small molecules (eg, TKIs)
| ||||||||||||||||||
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree