CALCITONIN GENE FAMILY OF PEPTIDES
Kenneth L. Becker
Beat Müller
Eric S. Nylén
Régis Cohen
Omega L. Silva
Jon C. White
Richard H. Snider JR.
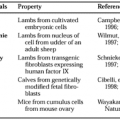
The calcitonin gene family consists of five genes (CALC-I to CALC-V)that are located on chromosome 11 (CALC-I, CALC-II, CALC-III, and CALC-V) and on chromosome 12 (CALC-IV). In humans, the mRNAs arising from these genes generate multiple peptides, including calcitonin (CT), calcitonin gene–related peptide (CGRP), amylin, adrenomedullin (ADM), and various circulating precursor and derivative peptides, some of which may have biologic functions (Fig. 53-1).1,2,3,4 and 5
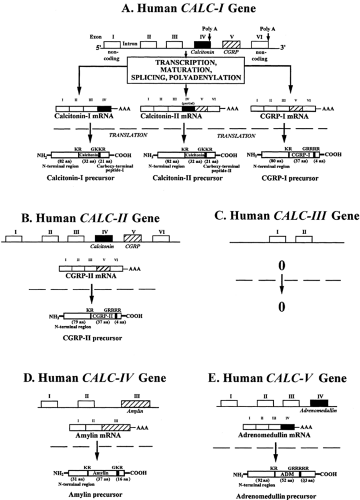 FIGURE 53-1. The human calcitonin (CT) gene family: organization of genes, mRNAs, and their hormone precursors. Based on their nucleotide sequence homologies, five genes belong to this family: CALC-I (CT/calcitonin gene–related peptide-I [CGRP-I]), CALC-II (CGRP-II), CALC-III, CALC-IV (amylin), and the CALC-V (adrenomedullin [ADM]) genes. Two structural features that are essential for full functional activity are conserved between the peptides: They contain two N-terminal cysteines that form a disulfide bridge resulting in an N-terminal loop and a C-terminal amide. A, The CALC-I primary transcript is processed into three different mRNAs: CT, CT-II, and CGRP-I mRNAs. The different products are generated by the inclusion or exclusion of exons by a mechanism termed splicing. Exons I–III are common for all mRNAs. Exon IV codes for CT, and exon V codes for CGRP-I. CT mRNA includes exons I + II + III + IV. CT-II mRNA includes exons I + II + III + IV (partial) + V + VI. CGRP-I mRNA is composed of exons I + II + III + V + VI. Each mRNA codes for a specific precursor. CT mRNA codes mainly for an N-terminal region, mature CT, and a specific C-terminal peptide (i.e., katacalcin, PDN-21, or calcitonin carboxy-terminal peptide-I [CCP-I]) that consists of 21 amino acids (aa). The N-terminal region includes a signal peptide of 25 amino acids and an N-terminal peptide of 57 amino acids (i.e.,aminoprocalcitonin [NProCT] or PAS-57). The CT-II precursor differs from the CT-I precursor by its specific C-terminal peptide, CCP-II. CCP-I differs from CCP-II by its last eight amino acids. CGRP-I mRNA codes for an N-terminal region, mature CGRP-I, and a cryptic peptide. The commitment of primary transcript in the different splicing pathways is determined, in part, by a tissue specificity. Although some overlap is seen, the CGRP-I mRNA is expressed mainly in nervous tissue, and CT mRNA is the major mRNA product in thyroid tissue and other tissues, whereas CT-II was found to be expressed in liver. B, The CALC-II gene codes only for a CGRP-II precursor. Its organization is similar to that of the CALC-I gene, containing 6 exons. Sequence homologies are important. Examination of the exon IV–like region of CALC-II indicates that CT mRNA is unlikely. Splicing at the site equivalent to the exon III–exon IV junction in human CT mRNA would result in a stop codon within the reading frame of the precursor polypeptide. Although CALC-II appears to be a pseudogene for CT, it is a structural gene for CGRP-II. The CGRP-II hormone differs from CGRP-I by three amino acids. C, The CALC-III gene contains only two exons. Their sequences have homologies with exon II and III of the CALC-I and CALC-II genes. The CALC-III gene does not seem to encode a CT- or CGRP-related peptide hormone and is probably a pseudogene that is not translated into a protein. D, The CALC-IV gene codes for a precursor containing the amylin peptide. This gene contains only three exons. The third exon codes for amylin. This 37-amino-acid peptide has marked homology with the CGRP peptides. The suggestion has been made that the CT and CGRP exons are derived from a primordial gene and that the different CT/CGRP/ADM/amylin genes have arisenby duplication and sequence-divergent events. E, The CALC-V gene is translated into ADM. This gene contains four exons. ADM is coded by the fourth exon. The amino-terminal peptides, encoded by exons II and III, also have some bioactivity.
Stay updated, free articles. Join our Telegram channel
Full access? Get Clinical Tree
 Get Clinical Tree app for offline access
Get Clinical Tree app for offline access

|