Acquired Immune Deficiency Syndrome
Learning Objectives
• Recognize the origin of human immunodeficiency virus 1 (HIV-1) and HIV-2
• Identify the HIV-1 clade predominant in most of the world
• Describe the structure of HIV-1
• Compare and contrast HIV-1 binding to lymphocytes and monocytes
• Explain the mechanisms involved in the fusion of HIV and the host cell membrane
• Explain the reverse transcription of HIV
• Understand the two-step process used to integrate viral deoxyribonucleic acid (DNA)
• Identify the process by which HIV exits host cells
• Discuss the roles of follicular dendritic cells in the pathogenesis of HIV
• List the possible mechanisms in the killing of CD4 cells
• Identify the possible mechanisms involved in CD8 dysfunction
• Identify HIV target cells in the brain
• Differentiate between the three states of HIV infection
• Compare and contrast HIV infection and acquired immune deficiency syndrome (AIDS)
• List the AIDS-defining medical conditions
• Understand the rationale for using highly active anti-retroviral therapy (HAART)
• Explain the mechanism of action of fusion inhibitors
• Compare and contrast nucleoside and non-nucleoside reverse transcriptase inhibitors
• Explain the mechanism of action of integrase inhibitors
Key Terms
Acquired immune deficiency syndrome (AIDS)
Capsid
Chemokine
Clade
Env
Follicular dendritic cells
Gag
Gp120/gp41
Iccosomes
Integrase
Matrix
Nef
Pol
Protease
Rev
Reverse transcriptase
Simian immunodeficiency virus (SIV)
Syncytia
Tat
Vif
Vpr
Vpu
Introduction
Human immunodeficiency virus (HIV) is closely related to simian immunodeficiency viruses (SIVs), which are found in equatorial Africa. SIVs are found in nine classes of monkeys and chimpanzees. Genetic sequencing strongly suggests that an SIV endemic to a chimpanzee subspecies (Pan troglodytes troglodytes) first crossed the species barrier in 1930. This virus is the ancestral HIV-1. In Gabon and Cameroon, the Sooty Mangabey monkey population is infected with a different SIV. Sometime between 1955 and 1972, the monkey SIV crossed the species barrier in several direct jumps to humans. The genome of this virus differs from HIV-1 by 30% to 40% and is designated HIV-2.
Subsequent studies linked HIV with the destruction of the immune system and acquired immune deficiency syndrome (AIDS). The syndrome is characterized by infections with unusual opportunistic pathogens (e.g., Pneumocystis jiroveci) and the development of aggressive sarcomas.
HIV-1 causes over 90% of human infections in the world. In the United States, 1.1 million individuals are infected with HIV-1, and 56,000 new cases are reported each year. HIV-2 is responsible for at least six epidemics in Africa. The World Health Organization (WHO) estimates that 4.3 million individuals are newly infected with HIV each year. Over 95% of these infections are occurring in the developing countries and Sub-Saharan Africa.
Human Immunodeficiency Virus Groups and Clades
According to the frequency of infections and the genetic diversity, HIV-1 is subdivided into groups and clades. HIV-1 is initially grouped into M (major), O (outlying), and N (new) groups. The HIV-1 M group has spread throughout the world. Groups N and O are restricted to West Equatorial Africa. Mutations within the M group, environmental pressure, and therapeutic selection have given rise to HIV clades. By definition, a clade is a virus subgroup that arose from a single common ancestor. The HIV-1 M group has nine major genetically diverse clades (A, B, C, D, F, G, H, J, and K). In the Western world, the B clade is the predominant virus. Transmission of HIV-1 B is rapid and associated with sex between men, intravenous drug use, and transmission of the virus from mothers to babies. HIV-1 A and C clades are found in southern and eastern Africa. Recombinant A-G and A-E clades also are emerging as infectious agents in Africa and Asia. In areas where subtypes C or A-E are common, heterosexual transmission is more frequent. Heterosexual transmission occurs because these clades replicate more efficiently in the Langerhans and dendritic cells of the vagina, and the virus is then shed into vaginal fluids. In the United States, the emerging trend in heterosexual HIV transmission has occurred because of the introduction of the A-E or C clade into North America.
Structure of the Human Immunodeficiency Virus
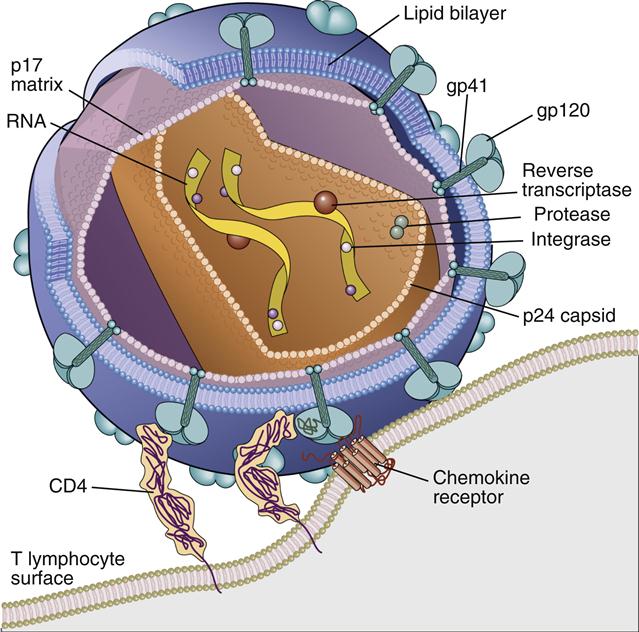
HIV virus comprises an inner core, a capsid, a matrix shell, and a host cell–derived outer membrane. The inner core contains two single strands of ribonucleic acid (RNA). One RNA strand, called the plus (+) or coding strand, contains viral genes that are transcribed. A second strand, the negative (–) strand, is complementary to the plus strand. The RNA and several proteins necessary for viral replication are encased in a cone-shaped capsid (Figure 26-1).
Additional protection for the capsid core is provided by an icosahedral shell or matrix. The shell anchors a lipid membrane to the outer surface of the virus. Protruding from the HIV membrane is the gp120 viral protein, which is anchored by a transmembrane gp41 protein. Three molecules of the gp120–gp41 aggregate to form a trimolecular structure necessary for docking to HIV target cells.
Human Immunodeficiency Virus Genes
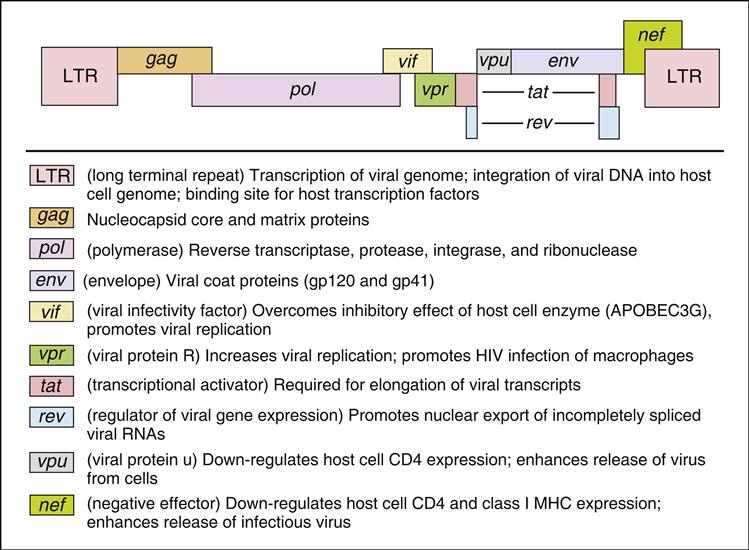
The HIV-1 genome codes for all the gene products necessary for viral replication. The genome is divided into structural genes (gag, pol, and env), regulatory genes (tat and rev) and accessory genes (vpr, nef, vpu, and vif). The organization of the HIV genome and the function of individual genes are shown in Figure 26-2.
Life Cycle of the Human Immunodeficiency Virus
The life cycle of HIV is divided into several stages, which include attachment, membrane fusion, uncoating, reverse transcription of viral RNA into DNA, and integration into the host cell genome. When HIV is integrated into the host cell DNA, it is called a latent provirus. Latency is characterized by the lack of viral gene expression. At some point in time, intracellular tat levels increase, and viral RNA is transcribed. After synthesis of multiple copies, the double-stranded RNA is packaged and exported from the cell.
Attachment
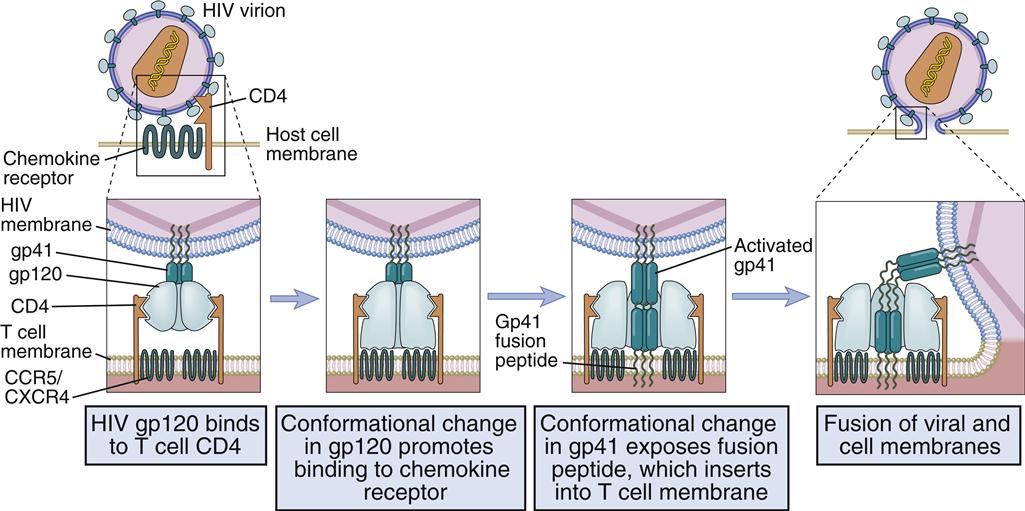
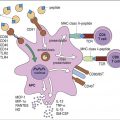
Different receptors are used to attach the virus gp120–gp41 to lymphocytes or monocytes. Attachment to T cells requires both CD4 and the chemokine receptor (CXCR4). Monocyte attachment points for HIV gp120–gp41 are a cell surface heparin molecule and a macrophage-specific chemokine (CCR5) receptor. Interaction with two receptors is necessary for the fusion of HIV with the host cell membrane (Figure 26-3).
Membrane Fusion
After interaction with gp120, the membrane attachment component (gp41) disassociates itself and undergoes a conformational change to become a long, cylindrical fusion protein, which bridges the divide between the virus and the cell membrane. After inserting itself into the host cell membrane, the gp41 fusion protein refolds itself using two specialized molecular H-1 and H-2 sequences called heptadrepeats. The H-1 and H-2 regions fold and form a six-helix fusion protein bundle, which brings the virus and host cell membrane into close contact (see Figure 26-3). Several six-helix fusion proteins form a narrow fusion pore in the membrane, which expands to allow viral entry into the cell.
After uncoating of the virus in the host cell cytoplasm, the nef gene is translated; it accelerates the endocytosis of the CD4 virus complex and downregulates the expression of class I molecules, which decreases the risk of the CD8-mediated destruction of infected cells.
Reverse Transcription (RT)
Pol gene products are critical for the synthesis and integration of viral DNA into the host genome and the generation of capsid proteins. Pol gene products include the HIV-1 reverse transcriptase, an integrase, and the late-phase protease. The pol gene proteins complex with RNase and vpr
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree