Surface Interactions Between T Cells and Antigen-Presenting Cells
Learning Objectives
• Discuss the clonal selection hypothesis and its relationship to the T cell receptor (TCR)
• Know the structure and functions of the α/β TCRs and γ/δ TCRs
• Define complementarity determining region
• Identify T cell subsets that interact with antigens presented on MHC class II molecules
• Understand the concept of somatic recombination in relationship to the TCR
• Explain the roles of RAG-1 and RAG-2 in the generation of TCR diversity
• Compare and contrast the junctional diversity and N- and P-nucleotide additions
• Recognize the components of the CD3 complex
• Differentiate between the signaling and nonsignaling components of the CD3 complex
• Identify TCR:MHC stabilizing molecules and their ligands
• Recognize the four critical co-stimulatory molecules and their roles in cell activation
• Compare the roles of B7-1 and B7-2 in the activation of T cell subsets
• Compare and contrast the roles of CD28 and CTLA-4 in T cell activation
• Understand how abatacept prevents T cell activation
• Understand how alefacept prevents T cell activation
• Recognize monoclonal antibodies that block TCR–HLA molecule interactions
• Explain the mechanism by which superantigens stimulate T cells
• Compare and contrast the pathophysiology of staphylococcal and streptococcal toxic shock syndrome
• Discuss the relationship between Vβ isoforms and toxic shock syndrome
• Recognize the immunologic defect in hyper-IgM syndrome
Key Terms
Abatacept
Alefacept
α/β T cell receptor (TCR)
B7 molecules
Complementarity determining regions (CDRs)
CD2
CD3 complex
CD4
CD8
CD11a
CD28
CD40L
Enterotoxin
γ/δ T cell receptor
Hyper-IgM syndrome
Immuno-receptor tyrosine-based activation motifs (ITAMs)
Junctional diversity
Monoclonal antibodies
Omenn syndrome
Somatic cell recombination
Superantigen
P-nucleotide additions
Introduction
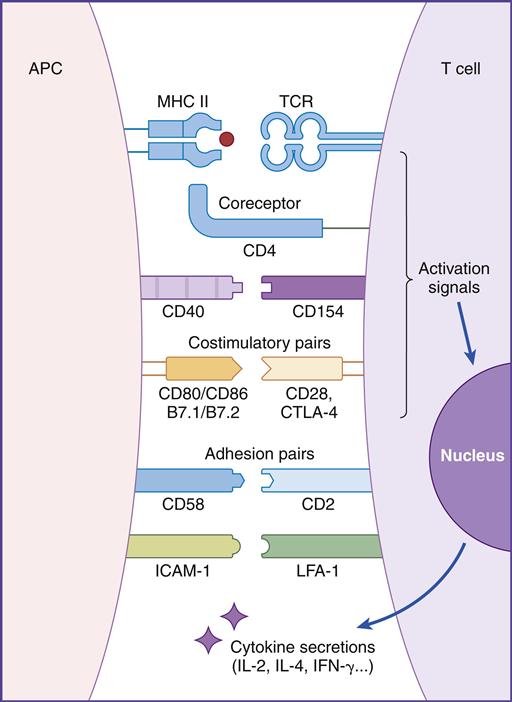
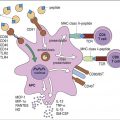
In this chapter, the discussion centers on the roles of receptor interaction in the activation or deactivation of T cells. Much like starting an automobile, several signals must be given in a defined sequence to override interlocking safety systems. For example, T cell activation requires three different steps: (1) The initial step entails interaction between the lymphocyte T cell receptor (TCR) and the antigen-loaded class II molecule. (2) In the second step, a CD4 molecule binds to class II molecules and stabilizes the TCR–class II complex. (3) In the final step of the activation sequence, the CD28 of the T cell interacts with a B7 molecule on the APC (antigen-presenting cell) (Figure 6-1).
If these three interactions do not occur, the T cell becomes nonfunctional for an extended period. In some cases, nonresponsive T cells become apoptotic and die.
T cell activation is central to the elicitation of an immune response. T cells act as effector cells or direct the immune response by influencing the activity of macrophages in the inflammatory response and of B cells in the synthesis of antibodies.
Clonal Selection Hypothesis
In the late 1950s, MacFarland Burnet proposed the clonal selection theory, which states that antigens, rather than lymphocytes, direct the immune response. The five basic tenets of the theory are (1) each lymphocyte has a unique receptor that binds to only one antigen; (2) the receptor must react with the antigen for cellular activation; (3) only selected clones of lymphocytes activated by antigen will divide (clonal expansion); (4) all of the daughter cells will express antigen receptors identical to the parent cell and (5) some of the daughter cells act as effector cells, and others become memory cells.
T Cell Receptors
The unique antigen binding receptor on T cells is called the T cell receptor (TCR). It is expressed on all T cells and exists in either α/β or γ/δ forms. The α/β TCR is expressed on peripheral blood T cells. γ/δ expressing T cells are rare in peripheral blood but are found in abundance in mucosal tissue. Each T cell has a homogeneous TCR population that recognizes a single antigen or epitope.
α/β T Cell Receptors
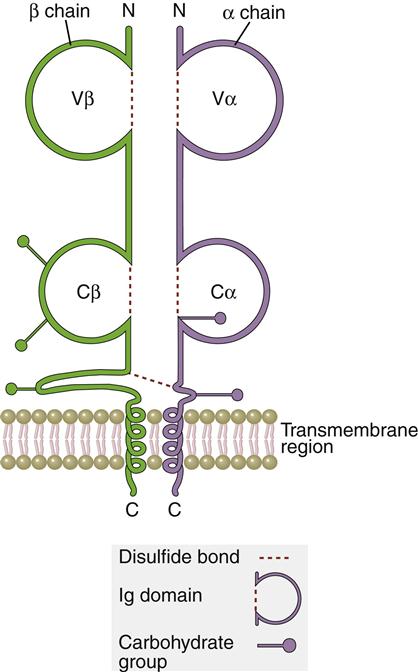
The TCR is a dual chain α/β heterodimer that consists of variable (V) and constant (C) regions. The structure of an α/β TCR is shown in Figure 6-2.
The V regions of the α/β-chains determine antigenic specificity by creating a three-dimensional pocket that recognizes immunogenic epitopes. The dual-chain V region pocket is created through the association of several different genes. The Vα pocket is the result of Vα genes joining with a junctional (Jα) gene. In the β-chain, a functional Vβ is created by linking Vβ, diversity (Dβ), and Jβ-domains.
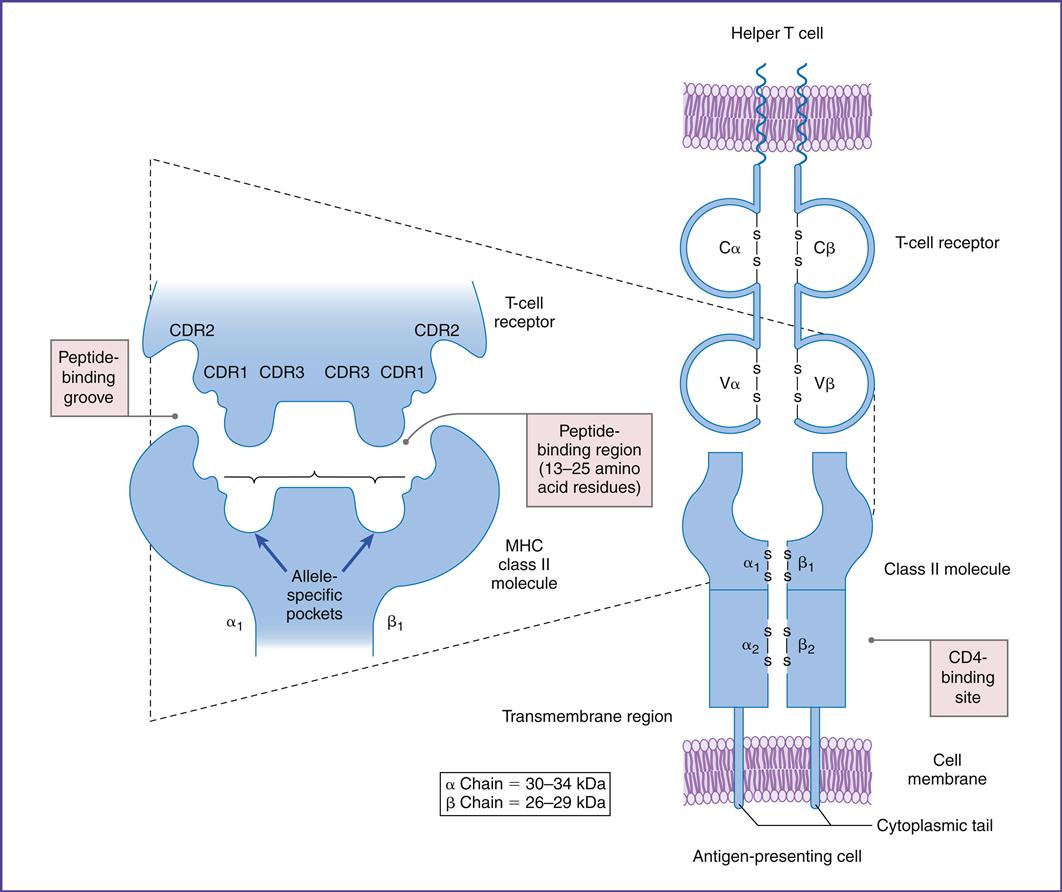
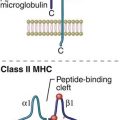
Complementarity determining regions (CDRs), which are hypervariable regions in the Vα/β-chains have actual contact with antigens. Class II molecules have six CDRs (three on each chain). The role of CDRs in binding to major histocompatibility complex (MHC) class II molecules is shown in Figure 6-3.
In the α-chains, CDR1 interacts with the N-terminal portion of the epitope. β-chain CDR1 reacts with the C-terminal portion of the peptide. CDR2s interact with the class II molecules but are not involved in antigen recognition. CDR3s are responsible for binding to the epitope presented by class II molecules. Most of the sequence variability responsible for TCR diversity is found in CDR3.
In addition to V regions, each TCR has an invariant constant (C) region. The C region consists of 140 to 180 hydrophilic amino acids and continues to a short hinge region with cysteine residues that link the α-chain and the β-chain A transmembrane anchor unit, which consists of highly charged hydrophobic amino acids, is connected to a short cytoplasmic region.
γ/δ T Cell Receptors
The Tγ/δ TCR structure is similar to the α/β TCRs. These T cells are only found in the mucosa and function as a bridge between innate and adaptive immunity. CD4γ/δ T cells recognize microbial phosphorylated molecules such as isopentylpyrophosphate (IDP) and dimethylallylpyrophosphate (DMAP). Direct interaction between these molecules and the γ/δ TCR activates the T cell. Antigen processing by APCs or presentation by MHC molecules is not required for activation. A vigorous γ/δ T cell response localizes the microbe in the mucosa until an adaptive response is mounted.
Origin of T Cell Receptor Diversity
In the generation of TCR diversity, two factors must be considered. First, each lymphocyte has a TCR that recognizes a single epitope. Second, 1013 different epitopes must be recognized. If a single lymphocyte TCR recognizes only one epitope, it follows that 1013 lymphocyte clones are present in each of the CD4 (Th1 and Th2) and CD8 (Tc1 and Tc2) populations.
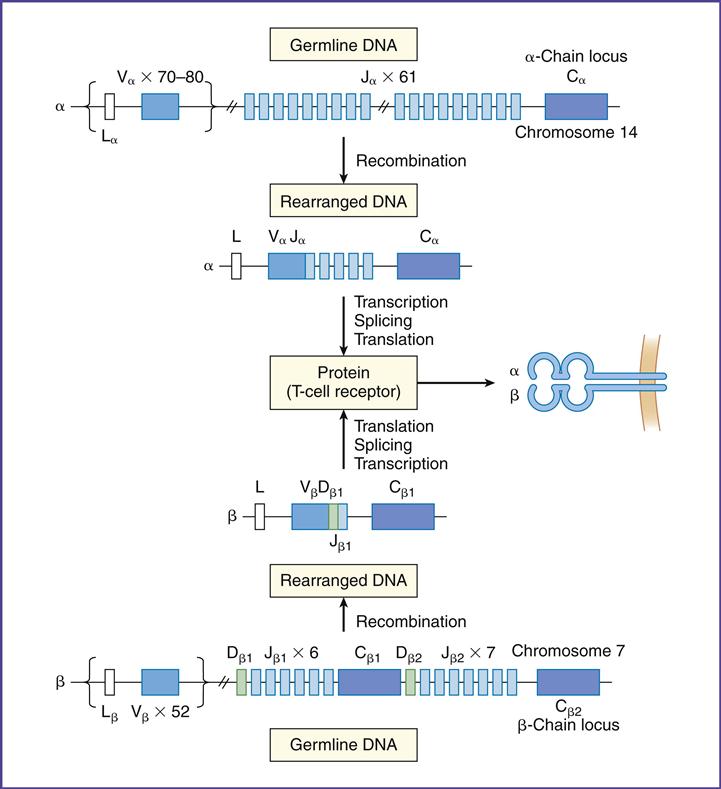
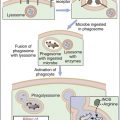
To create a repertoire of antigen-specific TCRs, alternative forms of genes present in somatic cells are rearranged in a process, called somatic cell recombination, by using RAG-1 and RAG-2 recombinase activating enzymes (Figure 6-4).
In the α-chain, 70 to 80 different Vα gene products can be linked to 61 different J-chains. Simple recombination in the α-chain can result in approximately 4.9 × 103 antigen-specific TCRs. β-chains have 52 variable gene segments, two diversity gene segments, and 13 J region genes. Recombination in the β-chain results in 1.3 × 103 unique TCRs.
Additional TCR diversity is created by alternate joining of Dβ-chains, junctional flexibility, and N- and P-nucleotide additions. Alternate joining of D domains creates 5 × 103 diverse TCRs. Adding six or more amino acids to the J domains (junctional diversity) creates more TCR diversity. Random insertion of palindromic sequences at single strand breaks (P-nucleotides) or nontemplated amino acids (N-nucleotides) into the TCR coding regions also occurs. Junctional diversity and P- and N-nucleotides generate an additional 1011 possible TCRs. The sum of the gene re-arrangements (4.9 × 103 + 1.3 × 103 + 5.3 × 10 3 + 1.0 × 1011) allows the immune system to respond to all known antigens.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree