Background
Breast carcinoma is the most common type of malignancy affecting women, aside from skin malignancy. It is the second most common cause of cancer deaths among women. Invasive breast carcinomas are a complex and heterogenous group of tumors, largely adenocarcinomas, with the majority falling into the category of invasive ductal carcinoma, not otherwise specified (NOS) . These tumors account for approximately 75% of all breast carcinomas. The second most common type of breast carcinoma is invasive lobular carcinoma, accounting for up to 15% of breast cancers. The terms “ductal” and “lobular” are not references to the cell of origin but rather how their associated in situ lesions affect lobular units. Ductal in situ lesions tend to expand and unfold lobular units, thus resembling ducts more than lobules. Lobular in situ lesions tend to expand but not distort the existing terminal duct lobular units (TDLUs). All breast carcinomas are thought to arise from the TDLU.
The remaining tumor types are of “special type” and are named according to their specific cytologic features and growth patterns. These tumors include tubular carcinoma, mucinous carcinoma, cribriform carcinoma, papillary carcinoma, metaplastic carcinoma, adenoid cystic carcinoma, apocrine carcinoma, neuroendocrine carcinoma, and secretory carcinoma, among others. Histopathologic features remain the standard for classification of breast carcinomas; however, the emergence of molecular diagnostics has allowed us to further classify tumors based on their molecular features.
Hormone Receptors and HER2
The three main receptors that are evaluated in breast carcinoma are estrogen receptor (ER), progesterone receptor (PR), and human epidermal receptor-2 (HER2). Immunohistochemistry (IHC) analysis of these three biomarkers, combined with Ki-67 IHC analysis for proliferative rate, can be used to approximate molecular subtyping. The different molecular subtypes of breast carcinoma have variable immunophenotypes and will be discussed later in this chapter. Hormone receptors and HER2 should be evaluated on all invasive carcinomas to determine the immunophenotype of the tumor.
Hormone receptor status is also a useful tool in patient management. ER and PR expression by breast cancer cells is a weak prognostic factor for outcomes but a strong predictive factor for response to endocrine therapy. Endocrine therapies include selective ER modulators (SERMs), aromatase inhibitors, and gonadotropin-releasing factor antagonists or refractory agonists. The most widely used endocrine therapy is tamoxifen, a type of SERM.
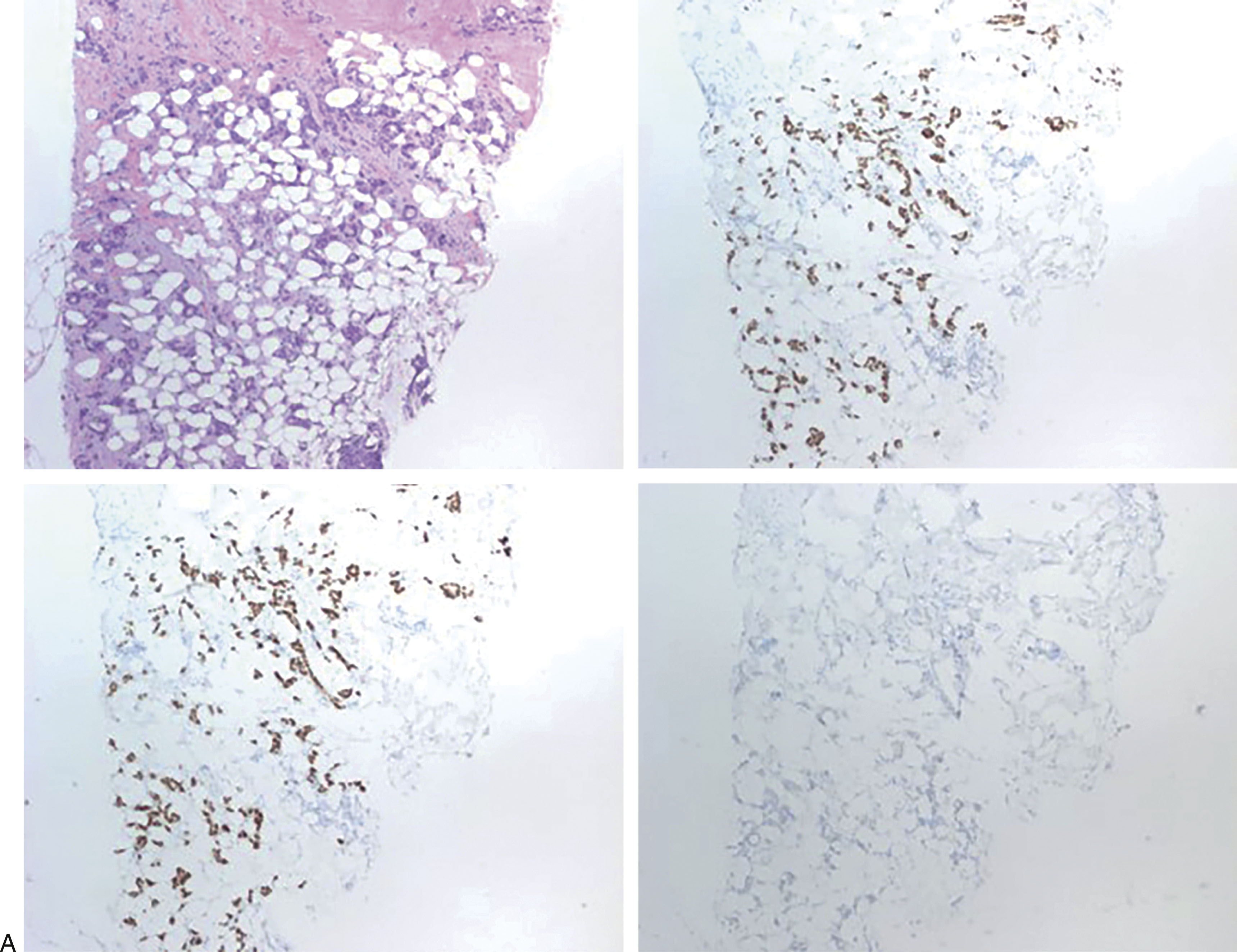
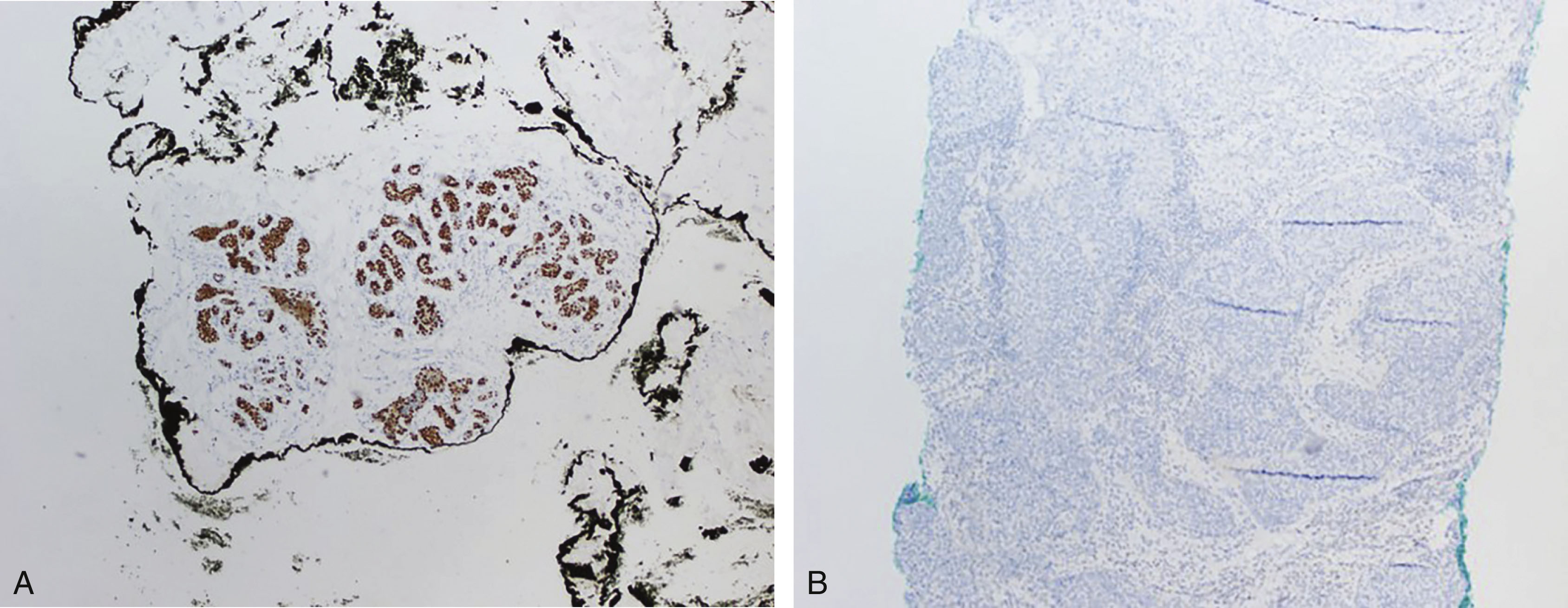
The best way to score ER and PR is still not standardized among pathology institutions. However, studies have shown that patients with as little as 1% ER positivity may benefit from endocrine therapy. Many institutions use a semiquantitative method for scoring ER and PR results, taking into account the percentage of tumor cells showing nuclear staining and the intensity (weak, moderate/intermediate, and strong intensity). The combination can be used to generate a score that can be used by clinicians in their treatment plans. The most widely used scoring system is the Allred scoring system. The most widely used cutoff percentages for ER and PR are 1% and 10% where less than 1% is a negative result, 1% to less than 10% is a low positive result, and ≥10% is a positive result. It should be noted that in the Allred system, a result of less than 1% with moderate or strong staining intensity is still considered a positive result, and patients may benefit from endocrine therapy. Hormone receptor markers are evaluated by staining the formalin-fixed paraffin embedded tumor samples using IHC and estimating the percentage of tumor cells staining and the intensity, under the microscope. This method of scoring remains relatively subjective, and many institutions have adopted the use of automated image analysis for the evaluation of ER and PR IHC results ( Fig. 10.1 ).
HER2 is another prognostic marker that should be evaluated in all breast cancers. It is a proto-oncogene present on chromosome 17q12. In HER2-positive breast carcinomas, HER2 is overexpressed, usually due to gene amplification. The HER2 protein is located on the surface of the tumor cells and makes a great target for therapy. HER2-targeted therapies include HER2-specific antibodies, such as trastuzumab, and small molecule inhibitors, such as the dual tyrosine kinase inhibitor lapatinib. Tumors that show HER2 overexpression or gene amplification have been shown to benefit greatly from treatment with trastuzumab and chemotherapy.
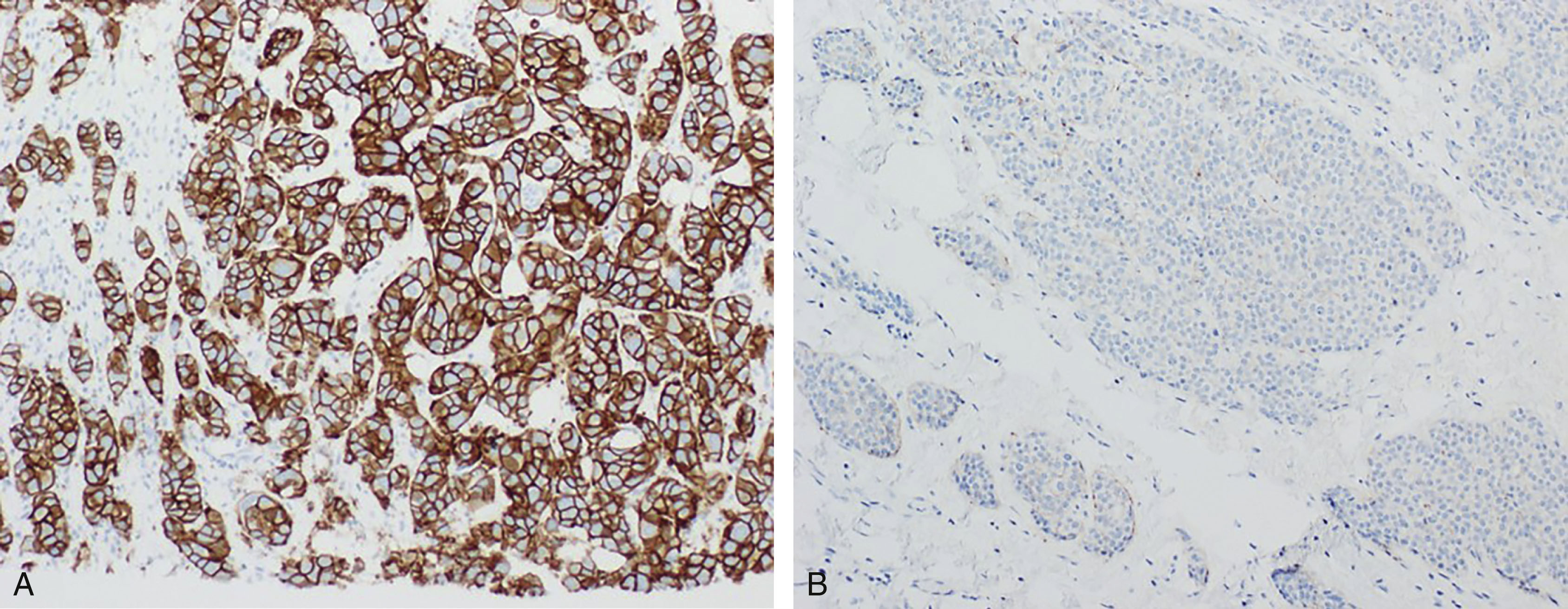
The most commonly used methods for evaluating HER2 status are with IHC to assess for overexpression and/or in situ hybridization (ISH) to assess for gene amplification. The scoring system for HER2 IHC analysis is well established through recommendations from the College of American Pathologists (CAP) and the American Society of Clinical Oncology (ASCO). Formalin-fixed paraffin-embedded tumor samples are stained using IHC for HER2 and evaluated under the microscope. Based on the staining pattern, the tumor is given an HER2 score of either 0 (negative), 1+ (negative), 2+ (equivocal), or 3+ (positive). A score of 0 is rendered when no staining is observed or membrane staining that is incomplete and is faint/barely perceptible and in ≤10% of tumor cells is present. A score of 1+ is rendered when there is incomplete membrane staining that is faint/barely perceptible and in greater than 10% of tumor cells. A score of 2+ is given when there is weak to moderate complete membrane staining observed in greater than 10% of tumor cells. Finally, a score of 3+ is given when there is circumferential membrane staining that is complete, intense, and in greater than 10% of tumor cells. Again, this method of scoring can be subjective, and many institutions use automated imaging software to analyze samples ( Fig. 10.2 ).
ISH analysis can be used in lieu of IHC; however, it is expensive and time-consuming. For cases that showed equivocal results by IHC, ISH should be performed on the sample to assess for the presence of gene amplification. The most widely used method for ISH analysis of HER2 gene amplification is using the dual probe method with a probe against the HER2 gene and the chromosome 17 centromere enumeration probe (CEP17). The number of HER2 signals and CEP17 signals per cell are evaluated and an HER2:CEP17 ratio is generated. The HER2:CEP17 ratio and HER2 copy number correspond to a “negative for gene amplification” or “positive for gene amplification” result with rare cases leading to an “equivocal” result. If an equivocal result is generated by ISH analysis, further testing is required.
The protein Ki-67 is used as a biomarker correlated with cellular proliferation, as it is present during all phases of the cell cycle but is absent from resting cells. IHC assays are used to determine a proliferative index using the Ki-67 antibody stain. Positive tumor cells are counted and divided by the total number of tumor cells evaluated. This process is tedious and can be subjective. However, there are now several image analysis systems to assist with this task. Ki-67 expression is correlated with grade, but among lower-grade tumors, Ki-67 can serve as an independent prognostic variable. Ki-67 is also used as a companion diagnostic for the CDK 4/6 inhibitor abemaciclib, as described below.
Molecular Classification of Breast Cancer
Gene expression profiling (GEP) studies have been able to identify several molecular subtypes for breast cancer through evaluation of DNA, mRNA, and proteins. The most well-understood subtypes are based on mRNA expression studies, and include luminal (A and B), HER2, and basal-like. A fifth molecular subtype of breast carcinoma, the normal-like breast cancer subtype, has been identified and closely resembles luminal A breast tumors. This only accounts for a very small percentage of all breast carcinomas.
These molecular subtypes show differences in clinical presentation, histopathologic features, response to therapy, prognosis, and molecular gene expression. Molecular classification of breast carcinoma does show reproducible correlation with prognosis, compared to traditional prognostic factors alone. The molecular subtypes can be correlated with ER status. The ER-positive subgroups include luminal A and luminal B subtypes and ER-negative subgroups include the HER2 and basal-like subtypes. While IHC staining as noted above can approximate these molecular subtypes, the agreement is not exact. Actual molecular subtyping is used in commercially available assays for prognosis, but a clinically reportable molecular subtype determination assay is not yet widely available.
Luminal Subtypes
The luminal A subtype of breast carcinomas accounts for ~55% of breast cancer and another ~15% of breast carcinomas fall into the luminal B subtype. The luminal subtype of breast cancer typically shows a high level of hormone receptor expression. Luminal A types of breast cancer tend to show higher levels of ER expression, and expression of genes regulated and activated by ER, when compared to luminal B types, but lower levels of proliferation-related gene expression. For this reason, Ki-67 IHC staining can be useful in approximating a luminal B phenotype. Luminal A subtypes are often PR positive by IHC, whereas luminal B subtypes are often PR negative or show low-level expression. HER2 is not typically expressed in luminal A cancers but can be expressed in up to 50% of luminal B cancers. The immunophenotype of luminal A tumors is often ER/PR positive and HER2 negative, whereas the immunophenotype of luminal B tumors is often ER/PR and HER2 positive ( Fig. 10.3 ).