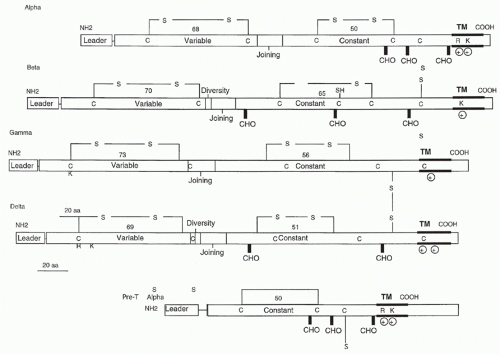
Subsequent subtractive cloning work rapidly identified two other candidate TCR complementary deoxyribonucleic acids (DNAs) identified as TCR-&agr;27,28 and TCR-&ggr;.29 It was quickly established that all antigen-specific helper or cytotoxic T cells expressed TCR-&agr;&bgr; heterodimers. Where TCR-&ggr; fit in remained a puzzle until work by Brenner et al.30 showed that it was expressed on a small (5% to 10%) subset of peripheral T cells together with another polypeptide, TCR-&dgr;. The nature of TCR-&dgr; remained unknown until it was discovered within the TCR-&agr; locus, between the V&agr; and J&agr; regions.31 Formal proof that the TCR-&agr; and -&bgr; subunits were sufficient to transfer antigen/MHC recognition from one T cell to another came from gene transfection experiments,32,33 and equivalent experiments have also been done with &ggr;&dgr; TCRs.34
containing two charged residues: an arginine and a lysine spaced identically to the TCR-&agr; transmembrane region. The cysteine in the connecting peptide is presumably what allows heterodimer formation with TCR-&bgr; and the similarity to TCR-&agr; in the transmembrane region is most likely to accommodate the CD3 polypeptides. Recently, Rossjohn and colleagues have presented a structure of the pre-T&agr; chain that is comprised of a single Ig-like domain that is distinct from the C domain of the TCR-&agr; chain; nevertheless, the mode of association between pre-T&agr; and TCR-&bgr; mirrored that mediated by the C&agr;-C&bgr; domains of the &agr;&bgr;TCR.39 The pre-TCR has a propensity to dimerize in solution, and the molecular envelope of the pre-TCR dimer correlated well with the observed head-to-tail pre-TCR dimer. This mode of pre-TCR dimerization enables the pre-T&agr; domain to interact with the V&bgr; domain through residues that are highly conserved across the V&bgr; and J&bgr; gene families, thus mimicking the interactions at the core of the &agr;&bgr; TCR’s V&agr;-V&bgr; interface. Disruption of this pre-T&agr;-V&bgr; dimer interface abrogates pre-TCR dimerization in solution and impaired pre-TCR expression on the cell surface. Their work suggests a mechanism of pre-TCR self-association that allows the pre-T&agr; chain to simultaneously “sample” the correct folding of both the V and C domains of any TCR &bgr;-chain, regardless of its ultimate specificity,39 which likely represents a critical checkpoint in T-cell development.
antigen or mitogenic stimulation of T cells64 and may play a role in T-cell activation as well.
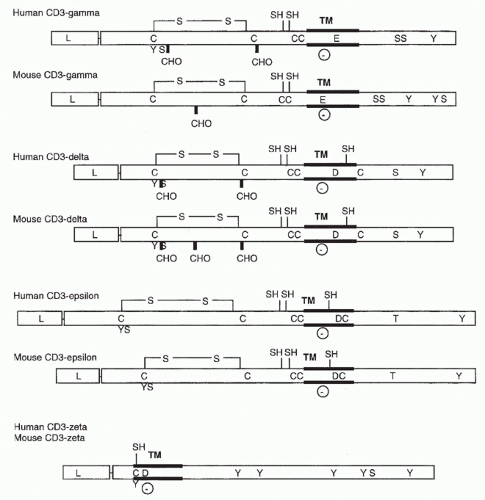 FIG. 11.2. Structural Features of the CD3 Molecules. As in Figure 11.1, transmembrane regions, carbohydrate addition sites, and cysteine residues are indicated. In addition, negatively charged transmembrane residues (D for aspartic acid and E for glutamic acid) and putative phosphorylation sites are shown. |
and &zgr;. In contrast, the TCR-&bgr; chain can assemble with any of the CD3 chains except the &zgr; chain. When the &zgr; chain was transfected with either &agr; or &bgr; chain genes, or any of the three CD3 chains, no pairwise interaction occurred.
receptor types are expressed in different ratios in different cells. These data may not be irreconcilable, because while the initial TCR/CD3 assembly may involve only one TCR, these may dimerize or multimerize later on the cell surface.50,75
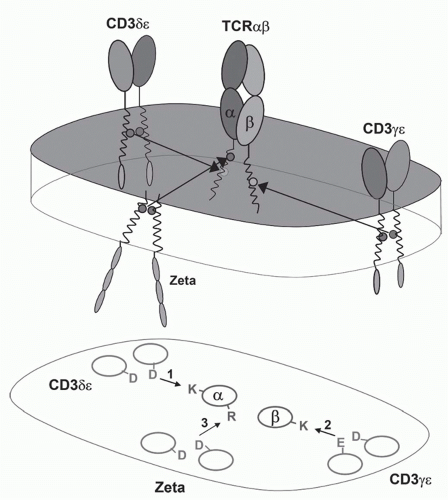 FIG. 11.3. A Model of the &agr;&bgr; T-Cell Receptor (TCR)-Cluster of Differentiation (CD)3 Complex. This shows the approximate positions of the &agr;&bgr; TCR chains and the CD3&agr;, &dgr;, &egr;, and &zgr; chains based on the membrane reconstitution and mutagenesis experiments of Call and Wucherpfennig.55 More recent experiments suggest that the CD3 polypeptides may be somewhat more clustered on the side of the TCR heterodimer.299 |
the functional diversity contributed by the J segment of the TCR (which constitutes a major portion of the complementarity-determining region [CDR]3 loop) makes an important contribution to antigen recognition (see the following).
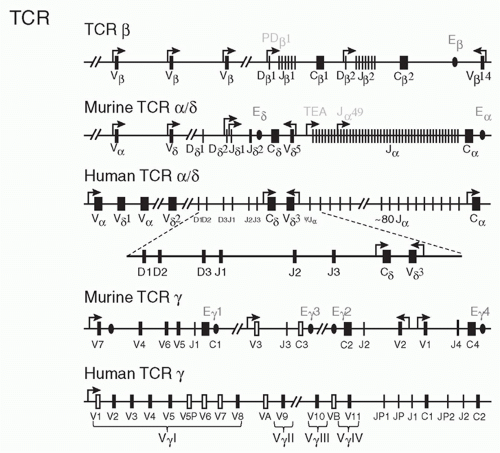
also express a V gene, V&bgr;17, that is not expressed in other strains of mice. Deletion of about half of the V genes (in SJL, C57BR and C57L mice) does not seem to have any particular effect on the ability of these mice to mount immune responses whereas mice that have deleted the J&bgr;2 cluster show impaired responses.94
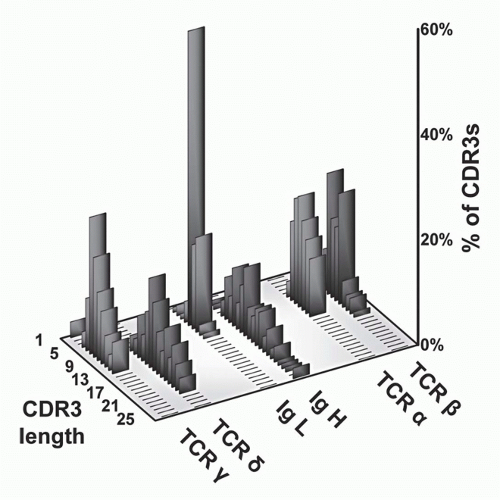 FIG. 11.5. Complementarity-Determining Region (CDR)3 Length Analysis of T-Cell Receptor (TCR) Polypeptides versus Immunoglobulin (Ig) Heavy and Light Chains. These data, modified from Rock et al.,129 show that whereas TCR-&agr; and -&bgr; CDR3 regions are relatively uniform with respect to each other, the other antigen receptor pairs show a marked asymmetry. Specifically, both Ig light chains (&kgr; and &lgr;) show very short CDR3s, as do TCR-&ggr; chains. In contrast, both IgH and TCR S TCRs are quite heterogeneous and tend to be longer. These data suggests that &ggr;&dgr; TCRs have a more antibody-like structure and binding properties. This has been borne out by subsequent analysis (see text). |
separately has little effect.127 In contrast to these results in thymocytes, deletion of “has” alone reduces transcription of one V&ggr; gene specifically in peripheral &ggr;&dgr; T cells. Thus, the two elements not only exhibit functional redundancy in thymocytes but also have unique functions in other settings.
and &bgr; chains of TCRs to contact both the MHC molecules and bound peptides on the same plane, as borne out by structural studies (see later section). In contrast, the CDR3s of Ig heavy chains are long and variable, whereas those of Ig light chains are short and constrained. This may reflect the fact that Igs recognize both very small molecules (eg, haptens) as well as very large ones (eg, proteins). Surprisingly, &ggr;&dgr; TCR CDR3 length distributions are similar to those of Igs in that the CDR3 lengths of TCR-&dgr; chains are long and variable, whereas those of the TCR &ggr; chains are short and constrained. Thus, on the basis of this measure of ligand recognition, one might expect &ggr;&dgr; TCRs to be more similar to Igs than to &agr;&bgr; TCRs. This has been validated in subsequent biochemical and structural studies (see later sections).
TABLE 11.1 Sequence Diversity in T-Cell Receptor and Immunoglobin Genes | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
putative proto-oncogenes that have been found translocated into the TCR-&agr;/&bgr; locus are the LIM domain-containing transcription factors Ttg-1151 and Ttg-2,152,153 which are involved in neural development; the helix-loop-helix proteins Lyl-1154 and Scl,155 which are involved in early hematopoietic development; and the homeobox gene Hox 11,156 which is normally active in the liver. How these particular translocations contribute to malignancy is unknown, but they presumably causes aberrations in gene expression that contribute to cell growth or escape from normal regulation. In patients with T-cell leukemia infected with the human T-cell lymphotrophic-I virus, there are large numbers of similar translocations; it is thought that human T-cell lymphotrophic-I itself is not directly leukemogenic but acts by causing aberrant rearrangements in the T cell that it infects, some of which become malignant.
TABLE 11.2 Chromosomal Locations of T-Cell Receptor, Immunoglobulin, and Related Loci in Mouse and Human | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
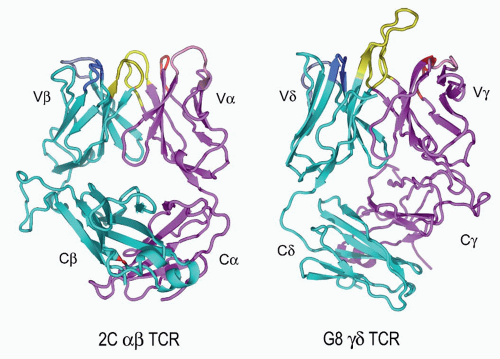 FIG. 11.6. Ribbon Diagrams of the T-Cell Receptor (TCR) Structures. This shows the structures of the 2C&agr;&bgr; TCR150 versus the G8&ggr;&dgr; TCR.153 The TCR-&bgr; and the -&ggr; chains are in cyan, and the TCR-&agr; and -&ggr; chains are in vermillion. The complementarity-determining regions (CDRs) of both are in yellow. The very long TCR-&dgr; CDR3 in G8, which binds the T10/T22 ligands, is very apparent here but is shorter in most other &ggr;&dgr; TCRs. Note the different C-region interactions with these TCRs and the deviations from the classic “&bgr; barrel” structure in both C&agr; and C&dgr;. The prominent C&bgr; loop to the left is also unusual and may mediate interactions with CD3 or other molecules on the T cell surface. (Figure courtesy of Dr. K.C. Garcia) |
In one structure,165 four out of seven N-linked sugars diffracted to high resolution, indicating that they are not free to move very much and thus are likely to play a structural role, particularly in C&agr;:C&bgr; interactions. This correlates with mutagenesis data indicating that certain C&agr; sugars cannot be eliminated without abolishing protein expression166 and the disordered state of a C&agr; domain in the structure of a TCR lacking glycosylation.164
There is significantly more contact between V&bgr; and C&bgr; and between V&agr; and C&agr; than in the equivalent regions of antibodies.
The geometry of the interaction of V&agr; and V&bgr; more closely resembles that of the CH3 domains of antibodies than VHVL.
Between the CDR3 loops of V&agr; and V&bgr;, there is a pocket that can (and does in at least one case165) accommodate a large side chain from the peptide bound to an MHC.
(or leucine zippered) molecules.179 One interesting variant that seems necessary for the stable expression of some class II MHC molecules in insect cells has been the addition of a covalent peptide to the N-terminus of the &bgr; chain.180
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree